1. INTRODUCTION
Climatic change is one of the most considerable challenges in today’s world. In the recent decades, the rise in temperature and frequent floods, especially in the midlatitude, decreased global crop productivity [1]. Plant breeders have been provided with an excellent opportunity to develop advanced cultivars with better desirable characteristics by using plant genetic resources [2]. Genetic material can be conserved for years together, by capturing and storing the plant genetic diversity in the plant genetic resources format, such as gene banks, in the repository and DNA library. The exciting advances in molecular genetics in recent years have provided specialists involved in the plant resources conservation with novel techniques for reputable and straightforward identification of diversity of the plant species [3]. Insufficient knowledge about the present genetic variations in plants and how to use it is one of the crucial motivations for conservation. Proper use of plant genetic resources, as well as the selection of high productivity and resistant variations, requires accurate identification of their accession [4].
Today, the central question for researchers is the translation of natural genetic polymorphism (genotype) into the phenotype and how plants adapt to the environmental changes. The 1,001 Genomes Project has addressed all these and related questions by providing high-resolution insight into global epigenetic and genetic polymorphisms in the model plant Arabidopsis thaliana [5] to help in the identification of specific genes with the help of genetic markers. The genetic marker concept is an adage, with Gregor Mendel in the late nineteenth century using a phenotype-based marker in his experiments. The limitations of these phenotypic-based markers subsequently led to the development of a DNA-based marker called a molecular marker. The molecular marker may or may not match with the genomic trait’s phenotypic expression, but it is stable and observable at all cell levels.
Additionally, the molecular markers are not confounded by ecological impacts. With the advent of DNA-based molecular markers, the practice of species identification technique has changed [6]. It has been developed into various techniques and used by numerous research groups throughout the world on a variety of plant species [7]. The main objective of this review is to provide the necessary information about the recently developed molecular marker technique and their application to genetic variability and variations in plant science for the conservation of plant resources.
2. ASSESSMENT OF GENETIC VARIATIONS IN PLANTS
Genetic diversity is one of the essential aspects of biological diversity that is significant for conservation programs [8,9]. The genetic variations in plants affect the higher level of biodiversity. The global population cannot adapt to and survive environmental changes without genetic diversity. Studies on the genetic variations are valuable for germplasm conservation, population, and variety identification and can identify alleles that might help the organism to cope with the changing environmental conditions [10]. Genetic variation assessment within and between plant populations is carried out using three techniques: (i) morphological, (ii) biochemical (allozymes), and (iii) DNA (molecular) marker techniques.
Morphological markers are simple and are based on visual inspection traits, such as growth habitat and flower color, and use inexpensive technologies, but they are time-consuming, labor-intensive, and need an expanse of land area and plant population for field experiments, which make them more expensive than molecular assessment. Morphological markers are highly dependent on environmental factors and are susceptible to phenotypic plasticity [2].
Biochemical markers are the allelic forms of enzymes (isozymes) that can be distinguished by a procedure of electrophoresis and specific staining techniques. These are the simplest and least expensive techniques to identify genetic variations between and among populations. Allozymes are allelic variants of enzymes encoded by structural genes and are usually a result of gene duplication. The resolution of the allozyme technique is low because only protein-coding regions of DNA can be assessed and only a small proportion of the changes in those regions will cause an evident change in the mobility of the protein [11]. These are codominant in nature and require a small amount of biological material for detection. Nevertheless, there are only a small number of isozyme markers available, and therefore the resolution of variation using biochemical markers is small or limited.
Molecular markers are the most widely used genetic markers, consisting of a wide range of DNA molecular markers, which can be used for the analysis of genetic variations. These markers are inherited for both dominance and codominance and may contain both expressed and nonexpressed sequences. These markers have been used and are best suited to study the genetic variations within populations [12]. DNA sequence variations in and among the plant species have also been found using different molecular markers [13].
3. ANALYSES OF GENETIC DIVERSITY BY MOLECULAR MARKERS
The systematic analysis of available molecular genetic in germplasm would be effective in resolving the genomic differentiation patterns that could be revealed by morphological-dependent taxonomic classifications. The molecular genetic variation data sets can render useful information on the allelic richness, population structure, and diversity parameters of germplasm, which can benefit the breeders to use genetic resources more effectively with fewer prebreeding activities for cultivar growth and improvement. Due to the quality and expeditiousness of data generated, germplasm characterization based on molecular markers has gained much importance nowadays [2].
A molecular marker is a DNA sequence on a locus in the genome of an organism at which the DNA genomic sequence varies among different individuals of a population. Molecular markers work by revealing variations (polymorphism) of the DNA sequences between different individuals in the population. These variations include insertions, deletions, point mutations, and translocations. An ideal biomarker has several preferred characteristics: (i) highly polymorphic and equally distributed throughout the genome; (ii) requires no previous knowledge of an organism’s genome; (iii) able to generate multiple, unique, and reliable markers and provide an adequate resolution; (iv) needs a small amount of start-up material and is simple, quick, and inexpensive; (v) does not possess pleiotropic or epistatic links to distant phenotypes.
It is difficult to obtain a molecular marker that meets the ideal criteria. A perfect molecular marker which fulfills most of the characters can be recognized depending on the nature of the study. Relying on the need, changes, and modifications in the techniques leads to the advances and second-generation molecular markers. Molecular markers are classified into three main categories or generations due to timeline of advancement in the technologies used: (i) hybridization-based markers, such as restriction fragment length polymorphism (RFLP); (ii) polymerase chain reaction (PCR)-based markers: random amplification of polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), and microsatellite or simple sequence repeat (SSR); and (iii) sequence-based markers: single nucleotide polymorphism (SNP).
The DNA-based molecular markers are commonly in use in many areas of plant science research, such as for genetic and phylogeny studies, as well as for the ecological, evolutionary, and taxonomic studies. These techniques are well developed and their advantages and limitations have been well defined [14,15]. A comparison of the most commonly used molecular markers is shown in Table 1, and the concept of the markers’ application is shown in Figure 1. Analysis of samples over a shorter period is possible by recent advances of high-throughput sequencing technologies.
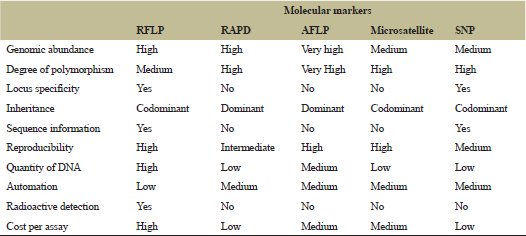 | Table 1: Comparison of the five most widely used molecular markers in plants. [Click here to view] |
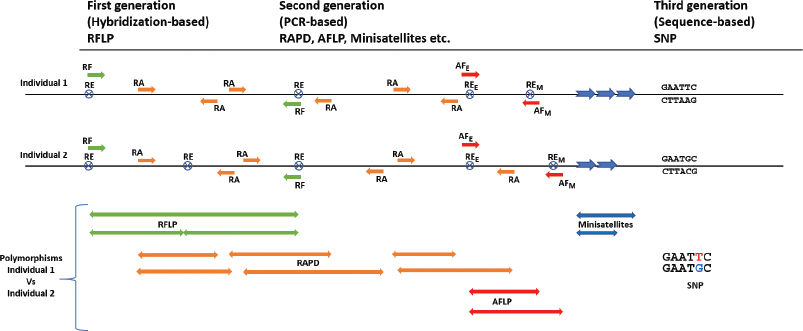 | Figure 1: Schematic representation of various molecular markers for the determination of polymorphisms in two hypothetical individuals. Arrows depict the direction of DNA fragments amplified or hybridized. RF: RFLP polymorphisms. RE: restriction enzyme sites; subscript E and M denote EcoR1 and Mse1, respectively, in the AFLP. RA: RAPD primers. AF: AFLP preselective and selective primers. Colored double-pointed arrows represent polymorphisms based on different molecular marker techniques. [Click here to view] |
3.1. Hybridization-Based Marker
Hybridization-based marker is named as such due to the process of identifying polymorphisms by hybridization of markers to the query DNA samples. In the beginning, RFLP had much power and was mostly used in the hybridization-based molecular marker. The RFLP marker was first used in 1975 to identify DNA polymorphism for genetic mapping of adenovirus serotype [16]. Later it was used for mapping human genes [17] and plant genomes [18]. In RFLP, variations in the characteristic pattern can be caused by any changes within a sequence of the DNA (point mutation), mutation between two sites (deletion and translocation), or mutation between the enzyme sites. RFLP is based on a restriction-hybridization technique where the DNA polymorphism of individuals can be detected by Southern blot hybridization of digested DNA to a chemically labeled DNA probe, resulting in the differential DNA fragment profile [4]. In some plants, such as wheat, however, low-frequency RFLP polymorphisms were observed, which can be attributed to the polyploidy nature of wheat and its large genome size [19].
RFLP markers are polymorphic, replicable, and codominantly inherited. They can be easily determined in the homozygous or heterozygous state of individuals. However, there are some limitations of the RFLP, such as being time-consuming and expensive and that it uses radioactive reagents and requires a large quantity of higher quality start-up material (DNA), rendering it less critical and rarely used and it is becoming obsolete [20]. These limitations led to the development of a new type of molecular markers called PCR-based molecular markers.
3.2. Polymerase Chain Reaction-Based Markers
The advent of studies in the field of molecular markers led to the development of PCR-based genetic markers for various purposes. PCR is a molecular biology technique used for amplifying a small amount of specific DNA region in vitro, enzymatically rather than in an organism itself. Generally, a short DNA strand up to 10 kb from a single or a part of a gene can be amplified using PCR. Several derivatives of PCR-based markers have been developed to suit the needs of various types of study.
3.2.1. Random amplified polymorphic DNA
The RAPD was the most straightforward PCR-based technique involved in the genetic variation analysis. The technique was developed independently by two different laboratories [21,22] and was known as RAPD and AP-PCR (arbitrary primed PCR), respectively. RAPD was the first to amplify the gene from species without DNA sequence information. This technique is based on the amplification of the random segments of the genomic DNA with the single primers of the arbitrary new sequence. The basic techniques of RAPD involve the following: (i) highly purified DNA; (ii) addition of single random primers; (iii) PCR; (iv) fragment separation by gel electrophoresis; (v) visualization of RAPD–PCR fragments; and (vi) fragment size determination, in comparison with a known molecular marker with the help of the gel analysis software.
RAPD was used to study genetic variations between plants of the same species. Genetic variations among populations of Capparis have been studied [23,24]. In the latter study, a total of 152 reproducible RAPD bands across the 36 individuals were amplified using 25 random primers. The result showed that the Farasan population had the highest level of genetic diversity (24.3%) and two populations [Khor Assos (5.9%) and Taif (4.6%)] had the lowest genetic diversity [23]. In another study, the genetic variations of five cultivars of Trigonella have been studied with 11 random primers, showing a total of 80 bands generated, of which 66 were polymorphic with a polymorphism of 82.50% [25].
.png) | Figure 2: RAPD marker profile of 10 landraces of V. umbellata generated by primer OPBB1 (Reprinted with permission [27]). [Click here to view] |
Genetic variation of Vigna umbellata was studied using the RAPD marker by primer OPBB1 (Fig. 2). The primer generated 987 bands, of which 719 were polymorphic with 70% polymorphism [26]. Helicanthus elastica (mango mistletoe) showed molecular differences in genomic DNA while growing on five different host trees when analyzed using four random RAPD primers [27]. RAPD was used to evaluate the genetic diversity among 20 Poincianella pyramidalis used in seed collection and conservation [28]. RAPD was also used together with intersimple sequence repeats (ISSRs) to study the genetic diversity of 18 Iranian populations of Nigella sativa, showing that these molecular markers can be useful for identifying and classifying plant species [29]. RAPD and SSR markers were also used to assess the genetic diversity of finger millet (Eleusine coracana), showing these molecular markers techniques are suitable for various plants, as well as for other organisms such as insects [30,31].
RAPD has been widely used because it is simple and cost-efficient and requires a small amount of DNA and no DNA probe and sequence information for primer designing. It is typically observed that, for assessment of genetic diversity, RAPD is usually used along with other molecular marker techniques [32]. However, there are few limitations of the RAPD marker, such as issues of less reproducibility and dominant inheritance. There is always a mismatch between primers in RAPD analysis. When the analysis using RAPD was conducted simultaneously with the other molecular marker methods, the other more advanced methods showed better genetic relationships between the analyzed samples [33].
3.2.2. Amplified fragment length polymorphism
Another PCR-based technique used to detect polymorphism is AFLP, which is based on the selective amplification by PCR of restriction fragments from a fully digested genomic DNA [34]. AFLP can be used to study genetic variations and is highly reproducible as it combines the reliability of RFLP with the sensitivity of PCR. This technique involves the following: (i) pure DNA from the samples to be analyzed; (ii) digestion of DNA with two restriction endonucleases mixture usually with a six-base-pair recognition site, such as EcoR1, and with a four-base-pair recognition site, such as Mse1; (iii) enzyme adapter ligation to the digested DNA; (iv) preamplification of restriction fragments with preselective primers with one extra base EcoR1+1 and Mse1+1; (v) selective amplification with fluorescently labeled primers with three extra bases EcoR1+3 and Mse1+3 (forward EcoR1+3 primer fluorescent labeled and reverse Mse1+1 unlabeled); and (vi) electrophoresis detection and fragment analysis. AFLP fragments are visualized either on an agarose gel with EtBr staining or on denaturing polyacrylamide gels with silver staining or automatic DNA sequencers.
This technique is used to study the genetic diversity and polygenetic relationship not only between different species but also between closely related genotypes [35]. AFLP generates fragments of any genomic DNA without any prior knowledge of sequences and any sources. This molecular technique is still prevalently being used for plant genetic study until now. The genetic diversity assessment of Elymus tangutorum was studied using 14 primer combinations which show an average polymorphism of 91% per primer combination [36]. Genetic variations of 177 accessions of Panicum turgidum Forssk, representing 10 populations, showed that 100% polymorphism was analyzed using AFLP markers [37]. Genetic diversity within the Norwegian Rhodiola rosea germplasm collection was analyzed using five AFLP primers, which shows about 80% of polymorphism among the clones [38].
The genetic and morphological variations in Metroxylon sagu have been studied using AFLP, which showed a significant correlation between genetic and geographical distances [39]. Another example of the AFLP marker profile in M. sagu produced by seven pairs of EcoR1 and Mse1 primers from our research is shown in Figure 3. The genetic diversity of 11 cultivars of soybean from 18 AFLP primer combinations has been reported with 43% polymorphism [40]. AFLP markers have been used to determine the genetic diversity for the conservation of endangered species, such as Australian hidden beard heath (Leucopogon obtectus), spring pasqueflower (Pulsatilla vernalis) in Europe, and Chascolytrum bulbosum, a grass species that is native to Latin America [41–45]; germplasm collection [46]; and the diversity of plant-related pathogens [47].
AFLP analysis is more efficient than RAPD and is highly reproducible, reliable, and polymorphic. It requires no prior sequence information or probe generation and can generate multiple fragments all over the whole genome to check polymorphisms as compared to RAPD, RFLP, and microsatellite. Besides, compared to the other molecular marker techniques, the AFLP is preferred if the researcher’s main criteria is a technique with the most discriminatory power [48], and the data can be stored in a database like AmpliBASE MT [46]. AFLP markers sometimes display dominance and are labor-intensive and require several steps to produce results.
.png) | Figure 3: AFLP marker profile of M. sagu generated by seven primer pairs of EcoR1 and MSe1. [Click here to view] |
3.2.3 Microsatellites
Microsatellites or SSRs are polymorphic loci consisting of di-, tri-, or tetranucleotide tandem repeats present in DNA sequences and comprise primary short motif genes between one- and six-base-pair lengths [49]. It is believed that the variations in the number of tandemly repeated units are mainly due to spilt-strands, miss pairing during DNA replication, repair, and recombination [50]. The amplification of microsatellites for identification is conducted using the unique sequence of flanking regions as primers. Therefore, one single pair of PCR primer can produce different sized products of different lengths in every end in the same species. The PCR product is separated either by capillary or slab gel electrophoresis in an automated sequence.
The SSR technique has been used to start conserving endangered species, such as Calystegia soldanella [51], Tricyrtis ishiiana [52], Galium catalinense subspecies acrispum [53], and Primula reinni [54]. In another example, the genetic diversity of Pennisetum glaucum hybrids and their parental line was studied using 55 microsatellite markers. Out of 55 markers, 37 were amplified, producing 162 alleles with the highest polymorphic information content (PIC) of 0.837 [55]. Microsatellite markers have been used to study quantitative trait locus (QTL) mapping in A. thaliana and have been proven as efficient to control seed stock or cross between accession [56] (Fig. 4).
Recent advances employing new SSR markers are primers that were developed from apple and pear to study the genetic variations and relationship of hawthorn (Crataegus spp.) [57]. In another study, to develop new SSR markers for lentil, an enriched genomic library for adenine-cytosine (AC) and adenine-guanine (AG) repeats were constructed from the Lens culinaris cv Kafkas [58]. A candidate gene approach has been used to develop and identify the SSR markers of aquaporin genes for drought stress in plants and can be used in studying the level of gene expression of these genes in new species [59]. The expressed sequence tag (EST)_SSR markers were developed to assess the genetic diversity of 27 accessions of erect milkvetch (Astragalus adsurgens) using 51 random primer pairs, in which by using these newly developed markers, wild milkvetch accession showed higher genetic diversity [60]. Thirty-six SSR markers were developed for Ephedra sinica by transcriptome database mining for genetic monitoring [61].
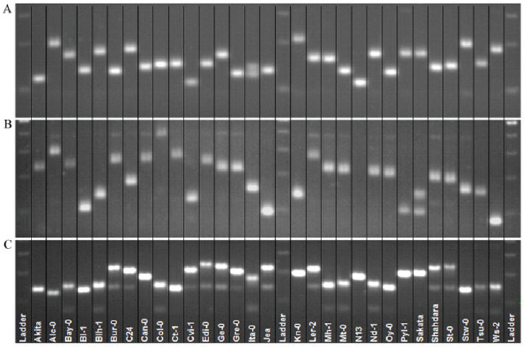 | Figure 4: Polymorphism obtained by three SSR markers is shown for 30 accessions of A. thaliana. (A) BSAT.024; (B) MSAT3.1; and (C) MSAT%.22 (Reprinted with permission [57]). [Click here to view] |
While the SSR polymorphisms are detected by a pair of primers that are unique to one locus in the genome and the primer pair amplifying the repeated portion of the microsatellites, another variant or subset of the SSR technique employs only a single primer for the study of polymorphisms. The technique is known as ISSR. Using the ISSR, one primer is used to amplify fragments between two identical but inversely oriented neighboring microsatellites. The ISSR is a dominant molecular marker, whereas the SSR markers are codominant; thus, they are able to detect both alleles in a heterozygous state. ISSR molecular markers are considered to have high marker efficiency, and SSR markers had extensive polymorphisms [62].
Microsatellites are considered to be a popular genetic marker. Unlike AFLP and RAPD, microsatellite markers show codominance. It is highly polymorphic, hypervariable, and is distributed throughout the genome and has a higher mutation rate than standard. However, microsatellite markers have few disadvantages as they require prior knowledge of the DNA sequence of the flanking region [63] and are not suitable for use across species [64]. The development of correctly functioning primers is expensive, time-consuming, and low throughput due to the difficulty for automation and data management [65].
3.3 Sequencing-Based Marker
3.3.1. Single nucleotide polymorphism
The SNP is a common type of variation in a DNA sequence among individuals of the same species. It has recently emerged as a new generation sequencing-based molecular marker, which can efficiently distinguish between homozygous and heterozygous alleles. The power in the SNP comes from the large number of loci that can be evaluated [66]. SNP provides the ultimate and reliable form of molecular markers as a single nucleotide base is the smallest unit of inheritance and can, therefore, provide an excellent marker density. SNPs are evolutionary consumed, which makes them less susceptible to the matter of homoplasy [67]. SNPs are suitable for high-performance automation, which allows effective genotyping of a greater number of samples [68]. The underlying protocol of SNP includes (i) sample reaction preparation with primers and templates, (ii) SNaPshot reactions using PCR, (iii) posttreatment of the products, and (iv) automated electrophoresis of samples and analysis of data.
In plants, SNPs can be designed from ESTs [69,70] and single-stranded pyrosequencing [71]. An SNP is used to construct the ultrahigh density genetic maps in plants and is used to determine genetic variations, especially in species with limited genetic diversity. The intra- and interpopulation diversities of broad beans (Vicia faba L.) were evaluated using a set of 768 SNP markers, of which 657 were amplified successfully and showed polymorphism [72]. The population genetic structure of castor beans (Ricinus communis) from genome-wide comparison was determined using SNPs, which shows the mixing of genotypes with low genetic variations, leading to the least worldwide geographic structuring of castor bean population [66]. In another study employing SNPs, 352 accessions of lentils (L. culinaris) originating from 54 diverse countries were reported to estimate the genetic diversity and genetic structure using 1,194 polymorphic SNPs [73]. SNPs markers were also used for genetic assessment and population structure analysis of 343 genotypes of spinach, showing that the genetic background in improved commercial F1 hybrids and several selected cultivars had a differently structured population from the United States Department of Agriculture (USDA) germplasm collection, which originated from different countries [74].
3.3.2. Next-generation sequencing (NGS)
The traditional DNA sequencing technologies could not meet the demand for in-depth sequencing information required in complicated genomic research. The advent of DNA sequencing technology has advanced the field of molecular biology [75]. NGS or second-generation sequencing technique has revolutionized genomic research by drastically decreasing the expenses and time requirement for sequencing using conventional sequencing methods based on the Sanger method (first-generation sequencing). NGS also revolutionized the study of variations among individuals in a population [2]. The NGS, followed by the third-generation sequencing approach, filled the gap of complicated genomic studies and made DNA sequencing partially crucial to the conservation biology [76]. The NGS and third-generation sequencing technologies available at present are 454 FLX (Roche), Illumina (formerly Solexa), supported oligonucleotides ligation and detection (SOLiD) (ThermoFisher), BGISEQ (BGI Genomics), single-molecule sequencing (Helicos), single-molecule real-time (SMRT) sequencing (Pacific Bioscience), PacBio RS II (Pacific Bioscience), and MinION (Oxford Nanopore) (Table 2).
The 454 FLX was the first NGS platform available, developed by 454 Life Science and is based on a pyrosequencing-based method that uses PCR to attain high-throughput parallel sequencing [77]. Meanwhile, the Illumina sequence platform was commercialized in 2006 and is based on a sequence-by-synthesis approach involving a basic library construction method with reversible fluorescence termination chemistry in the sequencing reaction, resulting in 35 bp reads [78]. The SOLiD system is another sequencing approach, which uses the ligation-based sequencing technique. The SOLiD4 platform has its origins in converting a cheap epifluorescence microscope as an automated nonelectrophoretic DNA sequencer [79]. A large set of SNPs was identified in a single walnut cultivar covering 98% of the physical map of the walnut genome when sequencing was conducted using the SOLiD4 platform [80].
 | Table 2. Next-generation sequencing platforms currently available. [Click here to view] |
A further advancement in the NGS technology resulted in a longer read high-throughput sequencing, known as third-generation sequencing. One example is Helicos true-single-molecule sequencing (tSMS), an exclusively unique DNA sequencing and genetic analysis approach, which is independent of PCR [81]. It offers a significant advantage over both traditional and NGS techniques. Helicos scientists have been able to increase the sample throughput of the system by five times (from 50 samples per run to 250 samples using the Helicos DNA Barcoding protocol, without losing precision or representational bias) [82].
The first commercial third-generation SMRT sequencer was PacBio RS II from Pacific Bioscience available in 2011 [83]. The data generated by the PacBio RS II have much better read lengths but are moderately lesser accurate than the second-generation platforms. To advance the read accuracy using this sequencing platform, a hybrid genome assembly approach called the “PacBio corrected Reads” algorithm was developed [84]. This algorithm of the hybrid genome assembly is based on the computational construction of precise longer consensus sequence by mapping high-accuracy second-generation NGS short reads to longer PacBio reads [85]. PacBio is preferred for polymorphism studies because of its long read length of over 10 kb on average, with some reads possibly reaching up to 60 kb. However, because of the PacBio high rates of random error in its single-pass reads, it is usually combined with another platform, such as Illumina [86]. This platform combination has also been used for SNPs and insertions/deletions (InDels) study in determining genetic variations between two varieties of the tea plant (Camellia sinensis) [87].
The MinION Nanopore sequencer is another new platform that can produce long sequencing reads on a palm-sized device that can be plugged into the USB port of a laptop [88–90]. Nanopore DNA sequencing is now an established sequencing platform that routinely achieves a read length of 50 kb and more and single-strand read accuracies of better than 92% [91]. The long read sequencing (LRS) systems of nanopore were used to assess low-coverage nanopore LRS for SNP genotyping in doubled haploid canola, showing a significant increase in the read length and improved alignment to the genome resulting in a more even representation of the genome [92]. Another option of high-throughput sequencing is the BGI Seq Complete Genomics nanoball technology by BGI Genomics. This method utilizes the amplification of small fragments of genomic DNA into DNA nanoballs by using the rolling circle replication method. The DNA sequencing using the BGI platform is comparable to the Illumina for use to generate high-quality data in DNA-related NGS applications. Using the BGISEQ-500 approaches (PE100 and PE75), more SNP events were identified in Arabidopsis compared to the analysis using Illumina HiSeq4000 [93].
NGS usually presents shorter read lengths, higher coverage, and different error profiles when compared to traditional sequencing data. Different kinds of software were specifically designed to cope with the NGS data. Many studies have reviewed the new software tools, methodology, advantages, and disadvantages of each NGS technology [94–96]. Since there are many different platforms of choice for conducting the sequencing, whether to elucidate the genome of any specific species or to further analyze the previously sequenced plant samples, for example, determination of species variability, the decision of which platform to use is usually decided by the number of reads and cost [94,97].
Comparing the preference and popularity of the molecular marker technologies that are now available in the market, the sequencing-based technologies are considered to be the preferred methods among researchers carrying out polymorphic and genetic studies. The current publication data showed that among the available technologies from the first-generation markers until the current third-generation markers based on DNA sequencing, all technologies are still being used and reported in prominent journal publications (Fig. 5).
The first-generation type of markers is still being employed now in 2020, albeit with reduced popularity. Based on the data obtained through the search engine lens.org [98,99], RFLP is still being used for polymorphic analysis, with the number decreasing from approximately 2,800 publications in 2009 to 1,200 in 2019, a 57% reduction in the number of publications in 10 years. The earlier second-generation type of markers, such as RAPD and AFLP, is less preferred with the number reduced to 70% over the same 10-year period. The minisatellites and microsatellites are the preferred second-generation molecular markers techniques among researchers, with the number of publications at almost a constant of 5,000 journal publications annually since 2009–2016, and which slightly reduced to around 3,300 in 2019. The preference for minisatellites and microsatellites is due to the more detailed information and accurate data sets of variations and polymorphisms that can be obtained using these methods compared to the earlier methods.
The current and most preferred method for studying polymorphisms is third-generation molecular marker analysis, employing the sequencing methods. The trend for using this sequencing approach has continuously been increasing from 2009, with both third-generation methods using SNPs and DNA sequence data, resulting in more than 9,000 publications in 2009 to the highest of 21,000 publications in 2015. From this trend, it can be observed that the SNPs analysis is the most preferred method for molecular markers analysis, most likely due to the advancement of technology for sample extraction, as well as the considerable reduction of the cost for conducting the analysis [100].
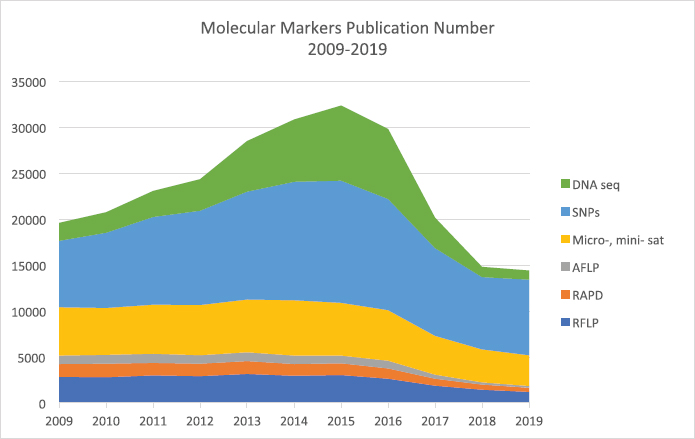 | Figure 5: The number of publications using various molecular marker techniques for polymorphism studies from 2009 to 2019. [Click here to view] |
4. CONCLUSION
This review summarizes some essential types of molecular markers that are developed among others to solve our major global issues of assessing plant responses to climate change. Molecular markers change the additional difficult phenotypical markers when investigating plant diversity that was used during the infancy of genetic research. The use of PCR-based DNA markers reviewed here shows that these markers have replaced hybridization-based markers, and this approach is increasingly widely used due to its simplicity and efficiency. These markers are used to determine the genetic relationships between closely related plant species. It also offers an insight into the mechanisms of somaclonal variation. Improvements in the DNA marker technique provide new possibilities of identification and analysis of commercially significant genes determining valuable, qualitative, and quantitative characters of plants. These markers are undoubtedly valuable tools for population genetics and plant breeding issue.
With the use of high-throughput molecular marker technologies ensuring speed and quality of data generated, it is possible to characterize a larger number of germplasms with limited time and resources. NGS reduced the cost and time required for sequencing the whole genome. Different types of molecular markers were developed and used for studying genetic variation and for the construction of genetic and physical maps. It is crucial to maintain, assess, and evaluate genetic diversity through these molecular marker techniques, as it provides a repository of adaptability to the environment and other changes as well. Since molecular genetics is a fast-growing field in science, new molecular marker techniques are likely to be developed further to overcome current limitations and provide advantages to researchers.
5. ACKNOWLEDGMENTS
This work was supported by the Ministry of Education Malaysia FRGS Grant (F07/FRGS/1611/2017) and UNIMAS Tun Openg Sago Chair Research Grant (F07/TOC/1743/2018). MN is the recipient of the Universiti Malaysia Sarawak Zamalah scholarship.
6. CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
7. REFERENCES
1. Kundzewicz ZW, Mata L, Arnell N, Doll P, Kabat P, Jiménez B, et al. Freshwater resources and their management. In: Parry ML, Canziani OF, Palutikof JP, van der Linden PJ, Hanson CE (eds.). Climate change 2007: impacts, adaptation and vulnerability contribution of working group II to the fourth assessment report of the intergovernmental panel on climate change. Cambridge University Press, Cambridge, UK, pp 173–210, 2007.
2. Govindaraj M, Vetriventhan M, Srinivasan M. Importance of genetic diversity assessment in crop plants and its recent advances: an overview of its analytical perspectives. Genet Res Int 2015;2015:14. CrossRef
3. Karp A, Seberg O, Buiatti M. Molecular techniques in the assessment of botanical diversity. Ann Bot 1996;78:143–9. CrossRef
4. Mondini L, Noorani A, Pagnotta M. Assessing plant genetic diversity by molecular tools. Diversity 2009;1:19–35. CrossRef
5. Kawakatsu T, Huang S-sC, Jupe F, Sasaki E, Schmitz RJ, Urich MA, et al. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 2016;166:492–505. CrossRef
6. Barcaccia G, Albertini E, Rosellini D, Tavoletti S, Veronesi F. Inheritance and mapping of 2n-egg production in diploid alfalfa. Genome 2000;43:528–37. CrossRef
7. Garrido-Cardenas JA, Mesa-Valle C, Manzano-Agugliaro F. Trends in plant research using molecular markers. Planta 2018;247:543–57. CrossRef
8. Gordon SP, López-Sepulcre A, Reznick DN. Predation-associated differences in sex linkage of wild guppy coloration. Evolution 2012;66:912–8. CrossRef
9. Kaljund K, Jaaska V. No loss of genetic diversity in small and isolated populations of Medicago sativa subsp. falcata. Biochem Syst Ecol 2010;38:510–20. CrossRef
10. Duran C, Appleby N, Edwards D, Batley J. Molecular genetic markers: discovery, applications, data storage and visualisation. Curr Bioinform 2009;4:16–27. CrossRef
11. Berta A, Sumich JL, Kovacs KM. Chapter 14 – population structure and dynamics. In: Berta A, Sumich JL, Kovacs KM (eds.). Marine mammals. 3rd edition, Academic Press, San Diego, CA, pp 533–95, 2015. CrossRef
12. Sica M, Gamba G, Montieri S, Gaudio L, Aceto S. ISSR markers show differentiation among Italian populations of Asparagus acutifolius L. BMC Genet 2005;6:17. CrossRef
13. Wang Y, Li XE, Li XD, Qi JJ, Sun P, Zhou LL. Analysis of genetic diversity of wild Rehmannia glutinosa by using RAPD and ISSR markers. Zhongguo Zhong Yao Za Zhi 2008;33:2591–5.
14. Agarwal M, Shrivastava N, Padh H. Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep 2008;27:617–31. CrossRef
15. Primmer CR. From conservation genetics to conservation genomics. Ann N Y Acad Sci 2009; 1162:357–68. CrossRef
16. Sambrook J, Williams J, Sharp PA, Grodzicker T. Physical mapping of temperature-sensitive mutations of adenoviruses. J Mol Biol 1975;97:369–90. CrossRef
17. Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 1980;32:314–31.
18. Helentjaris T, Slocum M, Wright S, Schaefer A, Nienhuis J. Construction of genetic linkage maps in maize and tomato using restriction fragment length polymorphisms. Theor Appl Genet 1986;72:761–9. CrossRef
19. Tanksley SD, Young ND, Paterson AH, Bonierbale MW. RFLP mapping in plant breeding: new tools for an old science. Biotechnology 1989;7:257–64. CrossRef
20. Cheriyedath S. Restriction Fragment Length Polymorphism (RFLP) Technique. News-Medical 2019; Accessed 09 March 2020, https://www.news-medical.net/life-sciences/Restriction-Fragment-Length-Polymorphism-(RFLP)-Technique.aspx.
21. Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res 1990;18:7213–8. CrossRef
22. Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 1990;18:6531–5. CrossRef
23. Abdel-Mawgood AL, Jakse J, Al-Doss AA, Assaeed AM. Genetic structure and diversity within and among six populations of Capparis decidua (Forssk.) Edgew. from Saudi Arabia. Afr J Biotechnol 2010;9:6256–63.
24. Khouildi S, Pagnotta MA, Tanzarella O, Ghorbel A, Porceddu E. Suitability of RAPD (random amplified polymorphic DNA) technique for estimating the genetic variation in natural genotypes of Tunisian and Italian caper (Capparis spinosa L.). Agricoltura Mediterr 2000;130:72–7.
25. Raja V, Wani UM, Kudesia R, John R. Estimation of genetic variability in Trigonella cultivars by RAPD analysis. Int J Biotechnol Res 2015;3:021–7.
26. Muthusamy S, Kanagarajan S, Ponnusamy S. Efficiency of RAPD and ISSR markers system in accessing genetic variation of rice bean (Vigna umbellata) landraces. Electron J Biotechnol 2008;11:32–41. CrossRef
27. Sunil Kumar KN, Maruthi KR, Alfarhan AH, Rajakrishnan R, Thomas J. Molecular fingerprinting of Helicanthus elastica (Desr.) danser growing on five different hosts by RAPD. Saudi J Biol Sci 2016;23:335–40. CrossRef
28. Belarmino KS, Rêgo MM, Bruno RLA, Medeiros GDA, Andrade AP, Rêgo ER. Genetic diversity in a Poincianella pyramidalis (Tul.) LP Queiroz population assessed by RAPD molecular markers. Genet Mol Res 2017;16:gmr16039663. CrossRef
29. Hosseini KorehKhosravi S, Masoumiasl A, Dehdari M. A comparative analysis of RAPD and ISSR markers for assessing genetic diversity in Iranian populations of Nigella sativa L. Cell Mol Biol (Noisy-le-Grand, France) 2018;64:52–9. CrossRef
30. Tello J, Forneck A. Use of DNA markers for grape phylloxera population and evolutionary genetics: from RAPDs to SSRs and beyond. Insects 2019;10:317. CrossRef
31. Prabhu KS, Das AB, Dikshit N. Assessment of genetic diversity in ragi [Eleusine coracana (L.) Gaertn] using morphological, RAPD and SSR markers. Z für Naturforsch C 2018;73:165–76. CrossRef
32. Bajpe SN, Bharathi TR, Marulasiddaswamy KM, Sampath Kumara KK, Prakash HS, Kini RK. Efficiency of RAPD, ISSR and ITS markers in detecting genetic variability among Salacia species sampled from the Western Ghats of Karnataka. Cell Mol Biol Lett 2018;45:931–41. CrossRef
33. Falakro K, Khiavi SJ. Assessment of genetic diversity and relationships among tea genotypes in Iran based on RAPD and ISSR markers. J Hortic Postharvest Res 2020;3:209–20.
34. Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 1995;23:4407–14. CrossRef
35. Althoff DM, Gitzendanner MA, Segraves KA. The utility of amplified fragment length polymorphisms in phylogenetics: a comparison of homology within and between genomes. Syst Biol 2007;56:477–84. CrossRef
36. Wu WD, Liu WH, Sun M, Zhou JQ, Liu W, Zhang CL, et al. Genetic diversity and structure of Elymus tangutorum accessions from western China as unraveled by AFLP markers. Hereditas 2019;156:8. CrossRef
37. Assaeed AM, Al-Faifi SA, Migdadi HM, El-Bana MI, Al Qarawi AA, Khan MA. Evaluation of genetic diversity of Panicum turgidum Forssk from Saudi Arabia. Saudi J Biol Sci 2018;25:123–9. CrossRef
38. Elameen A, Klemsdal SS, Dragland S, Fjellheim S, Rognli OA. Genetic diversity in a germplasm collection of roseroot (Rhodiola rosea) in Norway studied by AFLP. Biochem Syst Ecol 2008;36:706–15. CrossRef
39. Kjaer A, Barfod A, Lange C, Seberg O. Investigation of genetic and morphological variation in the Sago Palm (Metroxylon sagu; Arecaceae) in papua New Guinea. Ann Bot 2004;94:109–17. CrossRef
40. Zargar M, Romanova E, Trifonova A, Shmelkova E, Kezimana P. AFLP-analysis of genetic diversity in soybean [Glycine max (L.) Merr.] cultivars Russian and foreign selection. Agron Res 2017;15:2217–25.
41. da Silva L, Essi L, Welker C, Chies T. Assessing the genetic diversity and population structure of the endangered Chascolytrum bulbosum (Poaceae, Poeae) using AFLP markers. Biochem Syst Ecol 2016;68:236–42. CrossRef
42. Li X, Ding X, Chu B, Zhou Q, Ding G, Gu S. Genetic diversity analysis and conservation of the endangered Chinese endemic herb Dendrobium officinale Kimura et Migo (Orchidaceae) based on AFLP. Genetica 2008;133:159–66. CrossRef
43. Ronikier M. The use of AFLP markers in conservation genetics-a case study on Pulsatilla vernalis in the Polish lowlands. Cell Mol Biol Lett 2002;7:677–84.
44. van Ee BW, Jelinski N, Berry PE, Hipp AL. Phylogeny and biogeography of Croton alabamensis (Euphorbiaceae), a rare shrub from Texas and Alabama, using DNA sequence and AFLP data. Mol Ecol 2006;15:2735–51. CrossRef
45. Zawko G, Krauss SL, Dixon KW, Sivasithamparam K. Conservation genetics of the rare and endangered Leucopogon obtectus (Ericaceae). Mol Ecol 2001;10:2389–96. CrossRef
46. Majeed AA, Ahmed N, Rao KR, Ghousunnissa S, Kauser F, Bose B, et al. AmpliBASE MT™: a mycobacterium tuberculosis diversity knowledgebase. Bioinformatics 2004;20:989–92. CrossRef
47. Miller AM, Figueiredo JEF, Chaves CL, Ruas EA, Balbi-Peña MI, Colauto NB, et al. Genomic variability of Pantoea ananatis in maize white spot lesions assessed by AFLP markers. Genet Mol Res 2016;15:gmr15049452. CrossRef
48. Tabit FT. Advantages and limitations of potential methods for the analysis of bacteria in milk: a review. J Food Sci Technol 2015;53:42–9. CrossRef
49. Pradeep Reddy M, Sarla N, Siddiq EA. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 2002;128:9–17. CrossRef
50. Schlotterer C, Tautz D. Slippage synthesis of simple sequence DNA. Nucleic Acids Res 1992;20:211–5. CrossRef
51. Noda A, Nomura N, Mitsui Y, Setoguchi H. Isolation and characterisation of microsatellite loci in Calystegia soldanella (Convolvulaceae), an endangered coastal plant isolated in Lake Biwa, Japan. Conserv Genet 2009;10(4):1077–1079. CrossRef
52. Setoguchi H, Mitsui Y, Ikeda H, Nomura N, Tamura A. Development and characterization of microsatellite loci in the endangered Tricyrtis ishiiana (Convallariaceae), a local endemic plant in Japan. Conserv Genet 2009;10:705–7. CrossRef
53. McGlaughlin ME, Riley L, Helenurm K. Isolation of microsatellite loci from the endangered plant Galium catalinense subspecies acrispum (Rubiaceae). Mol Ecol Resour 2009;9:984–6. CrossRef
54. Yamamoto M, Handa Y, Aihara H, Setoguchi H. Development and characterization of 43 microsatellite markers for the critically endangered primrose Primula reinii using MiSeq sequencing. Plant Divers 2018;40:41–4. CrossRef
55. Tanwar H, Yashveer S, Sangwan VPS, Sharma S, Mor V. Molecular diversity analysis based on microsatellite markers in pearlmillet hybrids [Pennisetum glaucum (L.) R. Br.] and their parental lines. Indian J Ecol 2018;45(1):146–151.
56. Cosson P, Decroocq V, Revers F. Development and characterization of 96 microsatellite markers suitable for QTL mapping and accession control in an Arabidopsis core collection. Plant Methods 2014;10:2. CrossRef
57. Güney M, Kafkas S, Keles H, Aras S, Erci?li S. Characterization of hawthorn (Crataegus spp.) genotypes by SSR markers. Physiol Mol Biol Plants 2018;24:1221–30. CrossRef
58. Bak?r M, Kahraman A. Development of New SSR (Simple Sequence Repeat) Markers for Lentils (Lens culinaris Medik.) from Genomic Library Enriched with AG and AC Microsatellites. Biochem Genet 2019;57:338–53. CrossRef
59. Rabeh K, Gaboun F, Belkadi B, Filali-Maltouf A. In silico development of new SSRs primer for aquaporin linked to drought tolerance in plants. Plant Signal Behav 2018;13:e1536630. CrossRef
60. Gong W, Ma L, Gong P, Liu X, Wang Z, Zhao G. Development and application of EST-SSRs markers for analysis of genetic diversity in erect milkvetch (Astragalus adsurgens Pall.). Mol Biol Rep 2019;46:1323–6. CrossRef
61. Jiao SQ, Sun YQ, Zhang DX, Gao Q, Jin Y, Liu H, et al. Development of novel EST-SSR markers for Ephedra sinica (Ephedraceae) by transcriptome database mining. Appl Plant Sci 2019;7:e01212. CrossRef
62. Zhe L. Comparison of parents identification for tea variety based on SSR, SRAP and ISSR markers. J Tea Sci 2014;34(6):617–24.
63. Hollingsworth M. DNA fingerprinting in plants: principles, methods, and applications. In: Weising K, Nybom H, Wolff K, Kahl G. 2nd edition, 2005.: Boca Raton: CRC Press. $99·95 (paperback). 472 pp. Ann Bot 2006;97:476–7. CrossRef
64. Miah G, Rafii MY, Ismail MR, Puteh AB, Rahim HA, Islam KN, et al. A review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. Int J Mol Sci 2013;14:22499–528. CrossRef
65. Coates DJ, Byrne M. Genetic variation in plant populations: assessing cause and pattern. In: Henry RJ (ed.). Plant diversity and evolution: genotypic and phenotypic variation in higher plants. CABI Pub., Cambridge, MA, 2005.
66. Foster JT, Allan GJ, Chan AP, Rabinowicz PD, Ravel J, Jackson PJ, et al. Single nucleotide polymorphisms for assessing genetic diversity in castor bean (Ricinus communis). BMC Plant Biology 2010;10:13. CrossRef
67. Brumfield RT, Beerli P, Nickerson DA, Edwards SV. The utility of single nucleotide polymorphisms in inferences of population history. Trends Ecol Evol 2003;18:249–56. CrossRef
68. Tsuchihashi Z, Dracopoli NC. Progress in high throughput SNP genotyping methods. Pharmacogenomics J 2002;2:103. CrossRef
69. Coles ND, Coleman CE, Christensen SA, Jellen EN, Stevens MR, Bonifacio A, et al. Development and use of an expressed sequenced tag library in quinoa (Chenopodium quinoa Willd.) for the discovery of single nucleotide polymorphisms. Plant Sci 2005;168:439–47. CrossRef
70. Jiménez-Gómez JM, Maloof JN. Sequence diversity in three tomato species: SNPs, markers, and molecular evolution. BMC Plant Biol 2009;9:85. CrossRef
71. Pacey-Miller T, Henry R. Single-nucleotide polymorphism detection in plants using a single-stranded pyrosequencing protocol with a universal biotinylated primer. Anal Biochem 2003;317:166–70. CrossRef
72. Kaur S, Cogan N, Forster J, Paull J. Assessment of genetic diversity in faba bean based on single nucleotide polymorphism. Diversity 2014;6:88–101. CrossRef
73. Khazaei H, Caron CT, Fedoruk M, Diapari M, Vandenberg A, Coyne CJ, et al. Genetic diversity of cultivated lentil (Lens culinaris medik.) and its relation to the world's agro-ecological zones. Front Plant Sci 2016;7:1093. CrossRef
74. Shi A, Qin J, Mou B, Correll J, Weng Y, Brenner D, et al. Genetic diversity and population structure analysis of spinach by single-nucleotide polymorphisms identified through genotyping-by-sequencing. PLoS One 2017;12:e0188745-e. CrossRef
75. Gilbert W. DNA sequencing and gene structure Nobel lecture, 8 December 1980. Biosci Rep 1981;1:353–75. CrossRef
76. Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet 2008;9:387–402. CrossRef
77. Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005;437:376–80. CrossRef
78. Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev 2006;16:545–52. CrossRef
79. Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science (New York, NY) 2005;309:1728–32. CrossRef
80. You FM, Deal KR, Wang J, Britton MT, Fass JN, Lin D, et al. Genome-wide SNP discovery in walnut with an AGSNP pipeline updated for SNP discovery in allogamous organisms. BMC Genomics 2012;13:354. CrossRef
81. Schuster SC. Next-generation sequencing transforms today's biology. Nat Methods 2008;5:16–8. CrossRef
82. Blow N. DNA sequencing: generation next-next. Nat Methods 2008;5:267–74. CrossRef
83. Miyamoto M, Motooka D, Gotoh K, Imai T, Yoshitake K, Goto N, et al. Performance comparison of second- and third-generation sequencers using a bacterial genome with two chromosomes. BMC Genomics 2014;15:699. CrossRef
84. Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol 2012;30:693–700. CrossRef
85. Onda Y, Mochida K. Exploring genetic diversity in plants using high-throughput sequencing techniques. Curr Genomics 2016;17:358–67. CrossRef
86. Lin M, Qi X, Chen J, Sun L, Zhong Y, Fang J, et al. The complete chloroplast genome sequence of Actinidia arguta using the PacBio RS II platform. PloS One 2018;13:e0197393-e. CrossRef
87. Liu S, An Y, Tong W, Qin X, Samarina L, Guo R, et al. Characterization of genome-wide genetic variations between two varieties of tea plant (Camellia sinensis) and development of InDel markers for genetic research. BMC Genomics 2019;20:935. CrossRef
88. Jain M, Fiddes IT, Miga KH, Olsen HE, Paten B, Akeson M. Improved data analysis for the MinION nanopore sequencer. Nat Methods 2015;12:351–6. CrossRef
89. Ashton PM, Nair S, Dallman T, Rubino S, Rabsch W, Mwaigwisya S, et al. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol 2015;33:296–300. CrossRef
90. Check Hayden E. Pint-sized DNA sequencer impresses first users. Nature 2015;521:15–6. CrossRef
91. Jain M, Olsen HE, Paten B, Akeson M. The Oxford nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol 2016;17:239. CrossRef
92. Malmberg MM, Spangenberg GC, Daetwyler HD, Cogan NOI. Assessment of low-coverage nanopore long read sequencing for SNP genotyping in doubled haploid canola (Brassica napus L.). Sci Rep 2019;9:8688. CrossRef
93. Zhu FY, Chen MX, Ye NH, Qiao WM, Gao B, Law WK, et al. Comparative performance of the BGISEQ-500 and Illumina HiSeq4000 sequencing platforms for transcriptome analysis in plants. Plant Methods 2018;14:69. CrossRef
94. Kulski JK. Next-generation sequencing—an overview of the history, tools, and “Omic” applications. In: Kulski JK (ed.). Next generation sequencing–advances, applications and challenges. IntechOpen: London, UK, pp 3–60, 2016. CrossRef
95. Slatko BE, Gardner AF, Ausubel FM. Overview of next-generation sequencing technologies. Curr Protoc Mol Biol 2018;122:e59. CrossRef
96. Kumar KR, Cowley MJ, Davis RL. Next-generation sequencing and emerging technologies. Semin Thromb Hemost 2019;45:661–73. CrossRef
97. Ansorge WJ. Next generation DNA sequencing (II): techniques, applications. Next Gener Seq Appl 2016;1:1–10.
98. Jefferson OA, Jaffe A, Ashton D, Warren B, Koellhofer D, Dulleck U, et al. Mapping the global influence of published research on industry and innovation. Nat Biotechnol 2018;36:31–9. CrossRef
99. Jefferson R. Comment: turning science into social outcomes. Nature 2017;548:S8. CrossRef
100. Li F-W, Harkess A. A guide to sequence your favorite plant genomes. Appl Plant Sci 2018;6:e1030. CrossRef