Salt stress is the main constraint that limits agronomic output in several regions of the world. For the development of salt-tolerant crops, it is necessary to understand the salt-responsive proteins in halophytes. Myriostachya wightiana is a salt marsh that belongs to Poaceae. In this work, comparative proteomic studies were used to identify the proteins responsible for salt tolerance in the M. wightiana using two-dimensional electrophoresis and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Two protein spots 48 and 14 were over-expressed from salt-stressed M. wightiana and they were identified as chloroplast adenosine triphosphate (ATP) synthase β and glutamine-dependent nicotinamide adenine dinucleotide (NAD+) synthetase. The physicochemical characterization revealed that the ATP synthase β and glutamine-dependent NAD+ synthetase have 31714.6 and 66481.8 Da respective molecular weights. The secondary structure of ATP synthase β and Glutamine-dependent NAD+ synthetase has a greater percentage of random coils and alpha helix respectively. The modeled three-dimensional structures of ATP synthase β, Glutamine-dependent NAD+ synthetase have close similarities with 1fx0B and 3sytA. The functions of ATP synthase β and glutamine-dependent NAD+ synthetase were associated with the ATPase activity and non-covalent selective interaction with ATP correspondingly. This study provides information regarding salt-induced proteins to know the salt adaptation mechanisms for developing salt-resistant varieties.
Kumar MK, Sandeep BV, Sudhakar P. Identification and in-silico characterization of differentially expressed salt-induced proteins in the leaves of mangrove grass Myriostachya wightiana. J App Biol Biotech. 2020;8(05):48-58. DOI: https://dx.doi.org/10.7324/JABB.2020.80506
1. INTRODUCTION
Salinity is the main abiotic stress which influences all chief metabolic activities of a plant such as photosynthesis, protein synthesis, energy production, and lipid synthesis during the development [1]. Hence, salinity is regarded as the utmost severe restrictive factor for the productivity of crops and negatively influences the plant development [2]. Adaptation to salt stress is a complicated and multigenic response, which involves physiological, biochemical, and molecular pathways that can act collectively in the cell, tissue, and plant [3]. The salinity resistance of plants was not so far fully understand but explicated to some extent by stress adaptive effector mechanisms which include homeostasis of ions, biosynthesis of osmolytes, scavenging of toxic radicals, transport of water, management of long-distance response [4], and modifications in gene transcription which alters the expression patterns of proteins. There are many proteins to increase salt tolerance in plants by protecting themselves from cytotoxic ions by excluding through the plasma membrane or accumulating these ions in their storage vacuoles [5]. The improvement of salt-resistant plant varieties is the most important challenge in the present situation. The main problem in the development of salt-resistant plants is the lack of knowledge about the molecular mechanisms involved in salinity adaptations. Therefore, the studies related to the identification and characterization of the protein-based differential expression under salinity conditions are necessary. The identified proteins will provide a better way to develop salt-tolerant crop varieties by incorporating genes through genetic engineering.
Proteomics provides a platform to analyze the protein responses of plants to environmental stimuli and establish a link between the transcriptome and metabolome [6,7]. Proteomic approaches include extraction, purification, identification, and characterization. Protein identification can be carried out by gel-based separations followed by mass spectrometry (MS) [8]. Two-dimensional (2D) gel electrophoresis is an important technique for the separation of proteins and useful for determining the proteomic differences persuade by specific treatments or environmental alterations. Matrix-assisted laser desorption ionization-time of flight MS (MALDI-TOF MS) analysis commonly identifies the proteins based on the unique mass spectrum of peptide fragments through mass related bioinformatics tools and protein databases such as MASCOT and SEQUEST [9]. Bioinformatics gives a way to establish protein characterization such as protein structure, function, modifications, localization, and protein-protein interaction.
The biocomputation tools determine and characterize the protein’s function using the amino acid sequence. ProtParam is one of the protein analyzing tools in the ExPasy server which is used for computing various physicochemical parameters of a protein. Some bioinformatics programs have been developed for the prediction of proteins secondary structure. Self-optimized prediction method (SOPMA) is a recently developed tool to enhance the accuracy of the protein secondary structure prediction. The protein function is distinct by its structure; hence, approaches for determining protein structure from its sequence are gaining importance to integrate structural information into the annotation process [10]. Three-dimensional (3D) structure prediction is the main goal in the protein modeling from its amino acid sequence with a precision that is similar to the finest outcomes achieved experimentally [11]. Iterative Threading Assembly Refinement (I-TASSER) is a stratified protein modeling tool and it builds up possible structures by congregating fragments edited from the threading templates, where the biological perceptions of the modeled proteins are assumed by identical structures from the existed proteins in the databases [12].
Mangroves are classified as halophytes and they are found in estuaries and marine shorelines of tropical and subtropical tidal areas [13]. They divide into two groups such as true mangroves and mangroves associates. True mangroves specifically grow in intertidal regions while mangrove associates can grow in littoral and terrestrial habitats [14]. Studies on morphological, physiological, and biochemical alterations as an adaptation to salt stress in mangroves are inadequate to clarify the mechanism of salt adaptations. Recently some progress has been attained in perceptive the salt adaptation molecular mechanisms in mangroves, which are linked to the overexpression of salt-responsive proteins. Mangrove associates are perfect plants to study the salt-resistant mechanisms due to the transitional existences between terrestrial plants and mangroves. Myriostachya wightiana is a mangrove associate which is adapted to the two ecotypes, littoral, and terrestrial habitats.
M. wightiana is the imperative salt marsh in the family of Poaceae which, inhabitant laterally the muddy streams and channels in wetland mangrove swamps of Ganges Delta of India, Bangladesh and spreading into Myanmar, Malaysia, and Vietnam. In India, it is generally inhabited on the east coast of the Bay of Bengal. M. wightiana frequently inhabited with Acanthus ilicifolius, Nypa fruticans, and Porteresia coarctata. Saline water favors the growth and development of M. wightiana than freshwater due to the structural adaptive features such as a thick epidermis, sclerenchymatous vascular tissue, salt secreting pores, abundant metaxylem, and large phloem in the stem and leaves. The roots are adopted by the presence of thick cortex and lignified exodermis [15]. The literature review directs no research have happened on the M. wightiana. These characteristics make M. wightiana an ideal plant for the proteomic study. The current study focused on the identification and characterization of the salt-induced proteins from the leaves of M. wightiana.
Young and healthy seeds of M. wightiana were collected from the mangrove patch of Bhavanapadu which is located (Long: 18°33’ 52’’ to 18°32’ 11’’; N; Lat: 84°21’ 26’’ E to 84°18’ 22’’ E) on the North East of Andhra Pradesh, adjoining the Bay of Bengal, Tekkali, India. The collected seeds were shifted to sterile polyethylene zip bags and transported to the laboratory.
The field-collected healthy seeds of M. wightiana were first cleaned under tap water and then distilled water. After cleaning, the seeds were allowed to surface sterilization with 70% C2H5OH and 2% NaclO. Then, the sterilized seeds were soddened in distilled water overnight and kept for germination on a moisturized Whatman filter paper in a sterile Petri plate for 7 days. When the germination starts, the seedlings were shifted to pots loaded with autoclaved vermiculture. The pots were allowed to grow at 25°C 16 h photoperiod in a greenhouse and irrigated every day with ½ strength Hoagland solution. After 1 week, salinity treatment was given to test plants by irrigated with 500 mM NaCl solution whereas the control plants were irrigated only with water. Healthy and young leaves were collected from 4-week-old plants [Figure 1] to analyze the specific expression of salt-induced proteins.
 | Figure 1: Control and salt-treated Myriostachya wightiana. [Click here to view] |
Healthy and young surface-sterilized leaves (0.5 g) from the control and salt-stressed M. wightiana were finely pulverized separately by grinding in a motor and pestle with liquid nitrogen [16] and macerated with 2 ml homogenization buffer consists 50 mM Tris-HCl (pH 8.3), 0.5 M sucrose, 50 mM EDTA, 0.1 M KCl, 2 mM PMSF, and 0.1% 2-mercaptoethanol. The homogenate was allowed to centrifuge at 14,000 rpm for 10 min at 4°C. Finally, the supernatants from both control and test were kept at −20°C for further analysis. Lowry et al. [17] methodology was adopted to quantify the total protein concentration.
The protein separation from respective samples was done using 2DE. Isoelectric focusing (IEF) was used as the 1st dimension and it was done at 20°C using a 17 cm immobilized pH gradient (IPG) strip which has a 3 to 10 pH range. The IPG strips were rehydrated in 125 μl rehydration buffer (8 M urea, 2% CHAPS, 50 Mm dithiothreitol, and 0.2% biolyte) for 12 h by applying 50 V electricity and then the protein samples were loaded onto the strips. The IEF was performed in a stepwise manner: 150 V for 2 h, 300 V for 30 min, 1000 V for 30 min, 5000 V for 1.20 h, and 5000 V for 25 min. The focused strips were equilibrated 2 times for 30 min in 10 ml equilibration buffer (50 mM Tris-HCl, 6 M urea, 30% glycerol, 2% sodium dodecyl sulfate [SDS], and 100 mg dithiothreitol) with moderate shaking and subsequently additional equilibration for 15 min in equilibration buffer having 2.2% iodoacetamide instead of dithiothreitol. After equilibration, SDS-polyacrylamide gel electrophoresis was used as the second dimension and it was done using 10% separating gel. The vertical SDS gels were loaded with the equilibrated IPG strips. Finally, the gels were allowed to stain for overnight with 0.2% Coomassie Brilliant Blue G-250 and differential protein expression between control and treatment plants were measured by counting the number of protein spots on the gels.
The PMF of distinctively induced protein spots in the 2D gel of the test sample was analyzed with MALDI-TOF-MS [18]. Differentially displayed protein dots were extracted from the Coomassie-stained gel and the gel fragments were crushed and cleaned with 25 mM NH4HCO3 in 50% acetonitrile, then the gel fragments were subjected to dry and rehydration for overnight at 37°C in 25 mM NH4HCO3 with 1.0 mg of trypsin. Subsequently, the gel fragments were cleaned 3 times with distilled water, 50% acetonitrile and 5% trifluoroacetic acid at room temperature to obtain the proteins. 0.5 μl of extracted protein was placed on the matrix containing α-cyano-4-hydroxycinnamic acid made up of 50% acetonitrile and 5% trifluoroacetic acid. ABI 4800 MALDI TOF/TOF and GPS Explorer software (Applied Biosystems, Foster City, CA) was used for the collection of MS data and generation of peak lists from the raw data generated from the ABI 4800. The obtained mass spectrum was allowed to MASCOT (http://www.matrixscience.com) software for searching using sequence databases. The generated score by the MASCOT software indicates the chance of accurate identification and the value had to be as a minimum 50.
The identified protein sequences were recovered from National Center for Biotechnology Information (NCBI) in the FASTA format for Insilco analysis. The physicochemical characteristics of proteins such as molecular weight, isoelectric point (PI), number of positive and negative amino acids, extinction coefficient (EC) [19], instability index [20], aliphatic index [21], and grand average hydropathy (GRAVY) [22] were calculated with the ExPASy’s ProtParam server [23].
The secondary structural features of differentially expressed salt-induced protein sequences were calculated by subjecting the sequence to the SOPMA (https://npsa-prabi.ibcp.fr) server [24].
3D structural models for the identified proteins were developed by the Ab initio approach. I-TASSER Algorithms an online web server used to predict the structural models and functions of identified proteins [25]. ITASSER server predicts the protein structure depending on the multiple threading alignments built by LOMETS and iterative TASSER assembly simulations [26]. PROCHECK server was applied to generate (http://mordred.bioc.cam.ac.uk/rapper/rampage.php). Ramachandran plot was used for the accuracy validation of predicted 3D protein structural models and their stereochemical properties [27]. The quality of predicted 3D structures was confirmed by ERRAT (http://nihserver.mbi.ucla.edu/ERRATv2/) server.
The ITASSER server was used to predict the functional properties of modeled proteins basing on the global and local resemblances with the template proteins in Protein Data Bank (PDB). The template proteins were ranked depending on the global and local similarity scores and the annotations were subjected to top-scoring hits. The outputs of the server contain functional annotations of Gene Ontology (GO), Ligand binding sites, and Enzyme Commission numbers.
The salt-treated M. wightiana plant leaf has 11 ± 0.2% protein content per g fresh weight, whereas the control plant shows 13.2% protein. All the results were reported as mean ± SD obtained from three separate experiments. Due to NaCl treatment, total protein content was decreased in stressed plant leaves. Our findings agree with the results of earlier works like Nirjhar et al. [28] observed high protein concentration in freshwater-grown plants compared to salt-stressed plants. Zeynep et al. [29] stated that the protein concentrations are considerably decreased with increasing salt concentration and time intervals in all the tested cultivars of tomato. Results of Mohammad [30] have been revealed that the soluble protein content in rice is reduced with increased salinity. Joshi and Misra [31] observed that the percentage of protein in many plant tissues is decreased during drought or saline conditions, because of proteolysis and reduced protein synthesis. The present results are evident with Parida et al. [32] who found the increased activity of acid and alkaline protease during NaCl stress. The enhanced activity of proteases can act as an indication for salt stress and promote the reduction of protein levels in Bruguiera parviflora. Palma et al. [33] observed the irreversible oxidation of proteins by carbonylation. These oxidized proteins are selectively recognized and degenerate by proteases which lead to the loss of proteins.
2-DE provides an excellent method to compare the substantial quantitative expression of proteins between samples. In plants, salt stress triggers a huge number of stress-associated genes to produce a variety of functional proteins for countering stress. The comparative analysis of M. wightiana leaf proteome between the control and salt-stressed using 2DE with pH range 3 and 10 revealed a broad distribution of proteins from the PI range 4 to 9.5 in both control and salt-stressed plants. The maximum protein spots were found between the PI ranges of 4.5 and 7. Contrary to this, proteins which are fall in the acidic region, especially between the pH 4.5 and 5.5 showed a superior resolution. The highest percentage of protein variation was observed between the 6 KD and 34 KD markers regions in the gels of control and salt-stressed plants. M. wightiana comparative leaf proteome analysis significantly showed considerable changes in controlled and stressed conditions. A total of 81 protein spots were observed in a control plant whereas only 75 protein spots were found on the gel of salt-treated plant. The percentage of matching spots of the two gels was 90%. Fourteen spots showed quantitative and qualitative variations between the controlled and stressed plants. Among them, seven (3, 14, 17, 35, 48, 73, and 74) were up-regulated and nine were (27, 68, 69, 74, 75, 76, 77, 78, and 81) down-regulated in stressed plant. Two protein spots numbered as 14 and 48 were appeared very sharp and prominent. The molecular weight, PI values of 14, and 48 spots are 69 KD, 5.7, and 33 KD, 5.1 respectively. The prominent spots in the stressed plant were identified to understand the mechanism of adaptation against salinity. The 2D gel images control and salt-stressed M. wightiana were shown in Figure 2.
 | Figure 2: Two-dimensional (2D) gel images of Myriostachya wightiana comparative leaf proteomic expression under salt stress. (a) 2D gel image of control plant (b) 2D gel image of the salt-stressed plant. [Click here to view] |
The present results are evident with some previous reports which found that proteins involved in energy and metabolic regulation are changed under salt stress. In a cell, apart from the constitutive genes, several proteins participate in the functional execution and adaptation to environmental stimuli [34]. Peerzada et al. [35] stated that the treatment of salt stress could induce changes in protein expression. These differentially induced proteins are engaged with various cellular and physiological functions such as osmoregulation, redox homeostasis, ion balance, photosynthesis, energy requirements, and carbohydrate metabolism. Wang et al. [36] observed that Kandelia candel can sustain at 450 mM NaCl concentration possibly with the induced expression of proteins involved in light reactions that synthesize energy equivalents essential for the carbon fixation and other vital functions during salt stress. Page et al. [37] and Manna et al. [38] reported that the heat shock proteins were overexpressed with the treatments of cold and salt stress in tomato. Our findings show similarity with the results of Gomathi et al. [39] proposed that stress-induced high protein content possible by the specific expression of salt shock proteins having MW 15, 28, and 72 KDa in resistant varieties. Our results supported the findings of Majoul et al. [40] in wheat during heat stress. Production of energy depends on the adenosine triphosphate (ATP) synthase activity which is up-regulated in the present experiment as evident with the overexpression of spot 48, to make the energy needs for the encounter the ionic toxicity and other tolerance processes.
The unknown proteins are identified based on the spectrum of peptide masses which is known as PMF. For the construction of precise assignments, high-quality protein databases are immensely required to PMF [41]. MALDI-TOFMS of both the protein spots 48 and 14 from M. wightiana salt-stressed plant has a broad range of m/z values of 700–5000 Da. The mass spectrums of protein spots 48 and 14 were presented in Figures 3 and 4. For the protein identification, the obtained masses of peptide fragments were used to search in the NCBI database with MASCOT software. The up-regulated protein spot 48 was identified as chloroplast atpB gene product. PMF results of spot 48 show the highest score, that is, 89 with Oryza sativa chloroplast atpB gene product which is having Accession number gi|552857. The up-regulated protein spot 14 was identified as glutamine-dependent nicotinamide adenine dinucleotide (NAD+) synthetase. PMF results of spot 14 show the highest score, that is, 50 with O. sativa indica glutamine-dependent NAD+ synthetase gene product which is having Accession number gi|544604189. The protein identification results of spots 48 and 14 were shown in Table 1.
 | Figure 3: Matrix-assisted laser desorption ionization-time of flight mass spectrometry spectrof up-regulated protein spot (48) of salt-stressed Myriostachya wightiana leaf. [Click here to view] |
 | Figure 4: Matrix-assisted laser desorption ionization-time of flight mass spectrometry spectra of up-regulated protein spot (14) of salt-stressed Myriostachya wightiana leaf. [Click here to view] |
Table 1: Proteins identified from the 2DE gel spots of salt-stressed Myriostachya wightiana leaf.
| Spot no. | Max. homology with (Protein) | Best match organism | Expt. mw/Theor. mw (KD) | Expt. PI /Theor. PI | Score (MS/MS) | Accession no. |
|---|---|---|---|---|---|---|
| 48 | Chloroplast ATP synthase β subunit | Oryza sativa | 33/31.8 | 5.1/4.7 | 89 | gi|552857 |
| 14 | Glutamine dependent NAD(+) Synthetase | Oryza sativa | 69/66.5 | 5.7/5.86 | 50 | gi|544604189 |
ATP: Adenosine triphosphate, NAD: Nicotinamide adenine dinucleotide, PI: Isoelectric point, MS: Mass spectrometry
In the present results, it is observed that some of the up-regulated proteins having less molecular weight than the theoretical molecular weights. This might be from the degradation of proteins as a response to salt stress. Our results evident with the results of Lee et al. [42] observed the degradation of proteins which includes Rubisco large subunit, ATP synthase β subunit, FBP aldolase, and peroxiredoxin. Komatsu and Tanaka [43] reported that the rice seedlings have greater capability to synthesize ATP during oxidative stress to withstand in an anaerobic environment. Hence, the upregulation of ATP synthase is may be involved in the production of additional energy requirements to prevent the damage caused by salt stress. In salt-stressed Sorghum bicolor, the salt-induced proteins belong to the different functional groups which are involved in the salt stress adaptive mechanisms such as energy production and causes and signal transduction [44]. Juncheng et al. [45] found the upregulation ATP synthase β subunit in salt-stressed Medicago truncatula, and halophyte Halogeton glomeratus. Noctor et al. [46] stated that the synthesis of NAD is by the enzyme NAD synthetase. NAD plays an important role in various signaling and metabolic pathways which are connected with salt stress adaptation. Our result also implies that chloroplasts are one of the most influenced organelles inside cells by salt stress and chloroplast membrane proteins are highly susceptible to salt stress. To conclude, the differential proteomic studies in the Myriostachya leaf discloses that the salt stress affects the complex cellular network.
The primary sequences of the proteins ATP synthase β subunit and glutamine-dependent NAD+ synthetase of M. wightiana were retrieved from GenBank for homology, using BLAST P suite. The physicochemical characterization of ATP synthase β protein and glutamine-dependent NAD+ synthetase reveals that they have 294 and 593 amino acids with molecular weights of 31714.6 Da and 66481.8 Da, respectively. The results of physicochemical parameters shown in Table 2. The computed PI values of ATP synthase β subunit and glutamine-dependent NAD+ synthetase were 4.74 and 5.86, respectively. If the PI of a protein is <7, it is referred to as an acidic protein. At the PI, all the proteins show very least mobility. Hence, a buffer system development for the purification of protein is done using IEF. If all the Cys residues form cystines, the EC of ATP synthase β and glutamine-dependent NAD+ synthetase were 34505 M−1cm−1 and 94405 M−1cm−1, respectively. While if all the cysteines are reduced, the EC of ATP synthase β and glutamine-dependent NAD+ synthetase were 34380 M−1cm−1 and 93280 M−1cm−1 correspondingly. The EC of protein increases with the increasing concentration of amino acids Cys, Trp, and Tyr. The EC will be useful for the quantitative analysis protein based on UV spectral methods and the study of protein interactions in solutions with other proteins and ligands. The instability index of ATP synthase β and glutamine-dependent NAD+ synthetase was computed as 36.93 and 47.64 correspondingly. The instability index provides information regarding the stability of the protein in in-vitro conditions. If the protein instability index is <40, it is considered stable. Whereas, the instability index is computed as more than 40, the protein possibly unstable [20]. Rogers et al. [47] observed the in vitro half-life of proteins and reported that the instability index is more than 40, the protein shows <5 h half-life. If the protein has an instability index of <40, it shows more than 16 h half-life in in vivo conditions. The estimated half-life of ATP synthase β protein is 20 h in mammalian reticulocytes, 20 h in yeast. Whereas the half-life of glutamine-dependent NAD+ synthetase is estimated as 5.5 h in mammalian reticulocytes, and 3 min in yeast. Aliphatic index for the protein sequences of ATP synthase β and glutamine-dependent NAD+ synthetase was measured as 79.52 and 81.06, respectively. The aliphatic index is referred to as the concentration of aliphatic side chains present in the protein and it increases the thermal stability of globular proteins. The GRAVY indices of ATP synthase β and glutamine-dependent NAD+ synthetase were measured as −0.268 and −0.241. The negative value of the grand average of hydropathicity indicates that proteins are non-polar, hydrophilic (GRAVY typical value for hydrophilic protein is < −1), and better interaction of the protein with water. Our results show similarity with the findings of Kirchhoff et al. [48] who found ATP synthase CF1β subunit gi: 11467199 (5.31 PI) and (−0.078 gravy) located in the chloroplast thylakoid membrane.
Table 2: Physicochemical characterization of ATP β synthase subunit and Glutamine-dependent NAD+ synthetase.
| Parameter | ATP synthase β protein | Glutamine-dependent NAD(+) synthetase |
|---|---|---|
| Total no. of amino acids | 294 | 593 |
| Molecular weight | 31714.6 Da | 66481.8 Da |
| Theoretical PI | 4.7 | 5.86 |
| Negatively charged residues | 35 | 73 |
| Positively charged residues | 24 | 66 |
| Extinction coefficient | 34380 | 93280 |
| Instability index | 36.93 | 47.64 |
| Aliphatic index | 79.52 | 81.06 |
| GRAVY | -0.268 | -0.241 |
| Formula | C1379H2181N389O441S14 | C2937H4582N814O875S37 |
| Total number of atoms | 4404 | 9245 |
| Carbon | 1379 | 2937 |
| Hydrogen | 2181 | 4582 |
| Nitrogen | 389 | 814 |
| Oxygen | 441 | 875 |
| Sulfur | 14 | 37 |
ATP: Adenosine triphosphate, NAD: Nicotinamide adenine dinucleotide, PI: Isoelectric point, MS: Mass spectrometry.
SOPMA server was used to predict the secondary structure of proteins and it predicts the state of amino acid residues with an accuracy of 69.5%. In the secondary structure of ATP synthase β, the random coils and alpha-helix were found to be 39.12%, and 38.44% followed by extended strand 11.56% and Beta-turn 10.88%. The secondary structure of glutamine-dependent NAD+ synthetase has 45.53% Alpha helix, followed by 18.21% extended strand, 8.77% Beta-turn, and 27.49% Random coils. The secondary structural elements of ATP synthase β subunit and glutamine-dependent NAD+ synthetase were shown in Table 3. Secondary structure gives information regarding the amino acid existence, whether they are present in an alpha helix, β strand, or coil. The secondary structure of ATP synthase β and glutamine-dependent NAD+ synthetase reveals that these two proteins have a dominant percentage of random coils and α-helices while the extended strands and beta-turns exist in less percentage among all the secondary structural elements. The remaining secondary structural elements such as 310 helix, PI helix, ambiguous states, bend regions, and β bridges were not found in ATP synthase β and glutamine-dependent NAD+ synthetase. Secondary structure prediction of protein helps in understanding the hydrogen bonds present in the protein which further indicates the structural and functional efficiency [49]. The present observations show a strong relation to the functional properties of proteinaceous enzymes. The findings of this study evident with the observations of Vidhya et al. [50] reported that the greater concentrations of amino acid residues such as flexible glycine and hydrophobic proline enhance the percentage of random coils in protein secondary structure. Proline can interrupt ordered secondary structure by generating bends in the polypeptide chains. The stability and conservation levels of proteins are increase with the dominated percentages of random coils in structure [51]. The composition of proteins with a greater amount of hydrophobic amino acid residues especially in the transmembrane helices enhances the formation of complementary interactions with the hydrophobic lipid bilayer [52]. Bansal et al. [53] proposed that the functions of α-helical proteins are differentiated with their topologies which include signal recognition receptors, molecules, and ion transport across the membrane, translocation, and conservation of energy. The more extended structure of an α-helical protein may also help in sliding motion and their dynamics.
Table 3: Secondary structural elements of ATP synthase β subunit and glutamine-dependent NAD+ synthetase.
| Structural elements | ATP synthase β subunit | Glutamine-dependent NAD+ synthetase | ||
|---|---|---|---|---|
| Number of residues | Percentage of residues | Number of residues | Percentage of residues | |
| Alpha helix (Hh) | 113 | 38.44 | 270 | 45.53 |
| 310 helix (Gg) | 0 | 0.00 | 0 | 0.00 |
| PI helix (Ii) | 0 | 0.00 | 0 | 0.00 |
| Beta bridges (Bb) | 0 | 0.00 | 0 | 0.00 |
| Extended strands (Ee) | 34 | 11.56 | 108 | 18.21 |
| Beta turn (Tt) | 32 | 10.88 | 52 | 8.77 |
| Bend region (Ss) | 0 | 0.00 | 0 | 0.00 |
| Random coil (Cc) | 115 | 39.12 | 163 | 27.49 |
| Ambiguous states | 0 | 0.00 | 0 | 0.00 |
| Other states | 0 | 0.00 | 0 | 0.00 |
ATP: Adenosine triphosphate, NAD: Nicotinamide adenine dinucleotide.
3D structure of protein provides precise information regarding their interactions, stable conformation, and molecular functions. I-TASSER is an integrated computerized platform for the prediction of protein structure and function basing on the sequence. I-TASSER generates 3D atomic structural models to target protein sequence by threading for the possible folds using profile-profile alignments with the template structures which are selected from the PDB library. The target protein 3D structural models generated by comparing with the 3D models of selected template proteins. Then, the function of the protein was predicted. The proteins ATP synthase β and glutamine-dependent NAD+ synthetase have the best Z-score with the PDB ID: 1e79D and 3dlaA among the ten algorithms. The result of the structural alignment program reveals that the ATP synthase β and glutamine-dependent NAD+ synthetase protein sequences have close similarities with 1fx0B and 3sytA. Among the five models of ATP synthase β predicted by I-TASSER, the model with the best C-score (0.71) was selected with an estimated accuracy of 0.81 ± 0.09 (template modeling [TM]-Score) and 4.7 ± 3.1 A° (root-mean-squared deviation [RMSD]). The predicted best model of ATP synthase β protein was shown in Figure 5. Among the five models of glutamine-dependent NAD+ synthetase predicted by I-TASSER, the model with the best C-score (0.48) was selected with an estimated accuracy of 0.78 ± 0.10 (TM-Score) and 6.6 ± 4.0 A° (RMSD). The predicted best model of glutamine-dependent NAD+ synthetase was shown in Figure 6.
 | Figure 5: Three-dimensional structure of adenosine triphosphate synthase β subunit. [Click here to view] |
 | Figure 6: Three-dimensional structure of glutamine-dependent nicotinamide adenine dinucleotide+ synthetase. [Click here to view] |
The predicted models were validated basing on the geometric properties of the backbone conformations using Ramachandran plot analysis with the PROCHECK program. Ramachandran plot of the ATP synthase β shows [Figure 7], 72.3% residues are in most favored regions, 18.8% residues are in the allowed region, and 8.9% residues in the outlier region. Ramachandran plot of the glutamine-dependent NAD+ synthetase shows [Figure 8]. About 71.1% residues are in most favored regions, 18.6% residues are in the allowed region, and 10.3% residues in the outlier region. The present results confirm that most of the amino acids phi-psi distributions are constant with a right-handed α-helix and the models were stable and reliable. ERRAT program predicts overall quality for protein structural models by examining with the help of statistics of pairwise atomic interactions such as CC, CN, CO, NN, NO, and OO [54]. ERRAT server predicted that the overall quality factor of ATP synthase β and glutamine-dependent NAD+ synthetase 3D structures as 86.022 [Figure 9] and 84.906 [Figure 10], respectively. ERRAT confirms the quality of predicted 3D structures as the most reliable and within the accepted range. Consensus predictions suggested that selected proteins of M. wightiana contain ATP synthase and NAD+ synthetase family associated with various cellular activities.
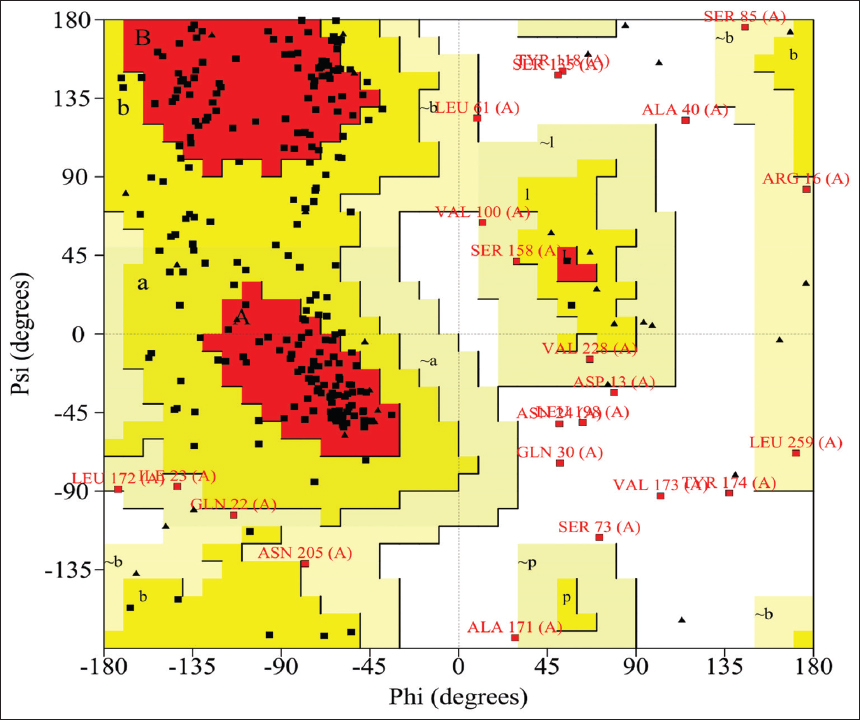 | Figure 7: Structural validation of adenosine triphosphate synthase β chain model by Ramachandran plot using Procheck server. [Click here to view] |
 | Figure 8: Structural validation of glutamine-dependent nicotinamide adenine dinucleotide+ synthetase protein model by Ramachandran plot using Procheck server. [Click here to view] |
 | Figure 9: Quality of predicted three-dimensional structure of adenosine triphosphate synthase β chain by ERRAT2 program. [Click here to view] |
 | Figure 10: Quality of predicted three-dimensional structure of glutamine-dependent nicotinamide adenine dinucleotide+ synthetase protein ERRAT2 program. [Click here to view] |
The functions of the query protein were predicted based on the functional analogs and the confidence score of the predicted 3D model. The protein modeling by the I-TASSER also predicts the GO basing on the functional homology score to the target protein. GO term explains the supposed function of the predicted structure. The modeled ATP synthase β molecular function, biological process, and cellular location were associated with GO:0008553, GO:0015991, and GO:0005886 with respective functions of hydrogen exporting ATPase activity, ATP hydrolysis coupled proton transport, and the protein is a component of the chloroplast thylakoid membrane. The modeled glutamine-dependent NAD+ synthetase molecular function, biological process, and cellular location were associated with GO:0005524, GO:0009435, and GO:0005886 with respective functions of non-covalently interacting with ATP, the formation of NAD from the nicotinic acid and the protein is a cellular component of biological membrane. The predicted numbers of GO terms and associated functions of ATP synthase β chain and glutamine-dependent NAD+ synthetase by I-TASSER server are listed in Table 4.
Table 4: Consensus prediction of ATP synthase β chain and glutamine-dependent NAD+ synthetase GO terms predicted by I TASSER.
| Functional parameter | ATP synthase β chain | Glutamine-dependent NAD+ synthetase | ||
|---|---|---|---|---|
| GO | GO Score | GO | GO Score | |
| Molecular function | GO:0008553 | 0.92 | GO:0005524 | 0.68 |
| GO:0005524 | 0.92 | GO:0003952 | 0.61 | |
| GO:0046961 | 0.92 | GO:0016810 | 0.36 | |
| GO:0046933 | 0.92 | GO:0016643 | 0.92 | |
| GO:0030228 | 0.41 | GO:0051540 | 0.36 | |
| GO:0005509 | 0.41 | GO:0016885 | 0.36 | |
| GO:0043499 | 0.41 | GO:0046872 | 0.33 | |
| GO:0042288 | 0.41 | -- | -- | |
| Biological process | GO:0015991 | 0.92 | GO:0009435 | 0.61 |
| GO:0006200 | 0.63 | GO:0040007 | 0.39 | |
| GO:0042777 | 0.62 | GO:0009084 | 0.36 | |
| GO:0006172 | 0.41 | GO:0006536 | 0.36 | |
| GO:0001525 | 0.41 | GO:0019319 | 0.36 | |
| GO:0006898 | 0.41 | -- | -- | |
| GO:0051453 | 0.41 | -- | -- | |
| GO:0006933 | 0.41 | -- | -- | |
| GO:0006629 | 0.41 | -- | -- | |
| Cellular component | GO:0005886 | 0.77 | GO:0005886 | 0.39 |
| GO:0009535 | 0.42 | GO:0005618 | 0.39 | |
| GO:0042645 | 0.41 | -- | -- | |
| GO:0009986 | 0.41 | -- | -- | |
| GO:0005754 | 0.38 | -- | -- | |
ATP: Adenosine triphosphate, NAD: Nicotinamide adenine dinucleotide, GO: Gene ontology.
Modeled ATP synthase β subunit protein representing EC 3.6.3.14 indicates the function of alpha/beta hydrolases fold type. The predicted possible ligand binding site residues of ATP synthase β were Y118, E142, R143, Y254, and Y266. Results were shown in Figure 11 and the possible binding ligand ANP (Phosphoaminophosphonic Acid-Adenylate Ester). Modeled glutamine-dependent NAD+ synthetase protein representing EC 6.3.5.1 deamido NAD+ L-glutamine amido-ligase. The predicted possible ligand binding site residues of glutamine-dependent NAD+ synthetase were K292, D293, I294, N296, E299, V331, Y332, M333, R400, and L412. Results were shown in Figure 12 and the possible binding ligand is ATP.
 | Figure 11: Ligand binding sites adenosine triphosphate synthase β chain. [Click here to view] |
 | Figure 12: Ligand binding sites of glutamine-dependent nicotinamide adenine dinucleotide+ synthetase. [Click here to view] |
In this study, comparative proteomic profiling in response to salinity in mangrove associate grass M. wightiana was performed by 2DE, MALDI-TOF, and Bioinformatics tools. Analysis of the leaf proteome revealed that there was a significant change in controlled and stressed conditions. The percentage of matching spots in the 2D gels of control and the salt-stressed plant was 90%. Fourteen spots showed quantitative and qualitative variations among the controlled and stressed plants. Among the 14, 2 prominently up-regulated proteins were identified in the leaves of salt-stressed M. wightiana as chloroplast atpB and glutamine-dependent NAD+ synthetase gene products. In silico studies of identified proteins revealed that they were involved in the energy requirements of M. wightiana by the enhanced production of ATP synthase β and maintained sufficient NAD+ levels by glutamine dependent NAD+ synthase under salt stress. The present results describe the high salinity tolerance methods of M. wightiana and explain the functional mechanisms involved in the salt-stress adaptation. This investigation can reflect the organization of cellular activities in M. wightiana under salinity stress, provides new perceptions in salt stress adaptation, and used as a source for the research works aimed to enhance salt resistance in rice and other plants.
The authors acknowledge the Department of Biotechnology, Andhra University, Visakhapatnam for the facilities made available.
Authors declared that they do not have any conflicts of interest.
University Grants Commission (UGC), Govt. of India for the financial support.
1. Parida AK, Das AB. Salt tolerance and salinity effect on plants: A review. Ecotoxicol Environ Saf 2005;60:324-49. [CrossRef]
2. Munns R, Tester M. Mechanism of salinity tolerance. Annu Rev Plant Biol 2008;59:651-81. [CrossRef]
3. Sairam RK, Tyagi A. Physiology and molecular biology of salinity stress tolerance in plants. Curr Sci 2004;86:407-21.
4. Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 2000;51:463-99. [CrossRef]
5. Hussain J, Rehman N, Khan AL, Hamayun M, Hussain SM, Shinwari ZK. Proximate and nutrients evaluation of selected vegetables species from Kohat Region Pakistan. Pak J Bot 2010;42:2847-55.
6. Cook D, Fowler S, Fiehn O, Thomashow MF. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci USA 2004;101:15243-8. [CrossRef]
7. Gray GR, Heath D. A global reorganization of the metabolome in Arabidopsis during cold acclimation is revealed by metabolic fingerprinting. Physiol Plant 2005;124:236-48. [CrossRef]
8. Agrawal GK, Sarkar A, Agrawal R, Ndimba BK, Tanou G, Dunn MJ, et al. Boosting the globalization of plant proteomics through INPPO: Current developments and future prospects. Proteomics 2012;12:359-68. [CrossRef]
9. Nesvizhskii AI, Keller A, Kolker E. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 2003;75:4646-58. [CrossRef]
10. Reeves GA. Genome and proteome annotation: Organization, interpretation and integration. J R Soc Interface 2009;6:129-47. [CrossRef]
11. Krieger E, Nabuurs SB, Vriend G. Homology modeling. Methods Biochem Anal 2003;44:509-23. [CrossRef]
12. Yang J, Roy A, Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 2013;29:2588-95. [CrossRef]
13. Mildred E, Mathias Botanical Garden. Mangal (Mangrove) World Vegetation. United States: University of California at Los Angeles; 2012.
14. Wang BS, Liang SC, Zhang WY. Mangrove flora of the world. Acta Bot Sin 2003a;45:644-53.
15. Rashid P, Ahmed A. Anatomical adaptation of Myriostachya wightiana Hook. f. to salt stress. Dhaka Univ J Biol Sci 2011;20:205-8. [CrossRef]
16. Giavalisco P, Nordhoff E, Lehrach H, Gobom J, Klose J. Extraction of proteins from plant tissues for two-dimensional electrophoresis analysis. Electrophoresis 2003;24:207-16. [CrossRef]
17. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with folin phenol reagent. J Biol Chem 1951;193:265-75.
18. Sheoran IS, Ross AR, Olson DJ, Sawhney VK. Proteomic analysis of tomato (Lycopersicon esculentum) pollen. J Exp Bot 2007;58:3525-35. [CrossRef]
19. Gill SC, Von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 1989;182:319-26. [CrossRef]
20. Guruprasad K, Reddy BV, Pandit MW. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Prot Eng 1990;4:155-64. [CrossRef]
21. Ikai A. Thermostability and aliphatic index of globular proteins. J Biochem 1980;88:1895-8.
22. Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol 1982;157:105-32. [CrossRef]
23. Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD. Protein identification and analysis tools on the ExPASy server. In: John M Walker, editors. The Proteomics Protocols Handbook. Totowa, New Jersey: Humana Press; 2005. p. 571-607. [CrossRef]
24. Geourjon C, Deléage G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci 1995;11:681-4. [CrossRef]
25. Zhang Y. Progress and challenges in protein structure prediction. Curr Opin Struct Biol 2008;18:342-8. [CrossRef]
26. Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc 2010;5:725-38. [CrossRef]
27. Lovell MR, Collins MW, Iverson GL, Field M, Maroon JC, Cantu R. Recovery from mild concussion in high school athletes. J Neurosurg 2003;98:296-301. [CrossRef]
28. Dasgupta N, Nandy P, Sengupta C, Das S. Protein and enzymes regulations towards salt tolerance of some Indian mangroves in relation to adaptation. Trees 2012;26:377-91. [CrossRef]
29. Zeynep BD, Koksal D, Hakan B, Ismail G. Effects of salt stress on pigment and total soluble protein contents of three different tomato cultivars. Afr J Agric Res 2010;5:2056-65.
30. Amirjani MR. Effect of NaCl on some physiological parameters of rice. EJBS 2010;3:6-16.
31. Joshi AJ, Misra H. Halophytic Grasses as Vital Components of Crop Halophytes. Sporobolus madraspatanus Borproc. Seminar on Sustainable Halophytes Utilization in the Mediterranean and Subtropical Dry Region. Osnabrueck: Osnabrueak University; 2000. p. 32.
32. Parida AK, Das AB, Mittra B, Mohanty P. Salt stress induced alterations in protein profile and protease activity in the mangrove Bruguiera parviflora. Z Naturforsch C J Biosci 2004;59c:408-14. [CrossRef]
33. Palma JM, Sandalio LM, Corpas FJ, Romero-puertas MC, Mccarthy I, Delrio LA. Plant proteases, protein degradation, and oxidative stress: Role of peroxisomes. Plant Physiol Biochem 2005;40:521-30. [CrossRef]
34. Kottapalli KR, Payton, Rakwal R, Agrawal GK, Shibato J, Burow M. Proteomic analysis of mature seed of four peanut cultivars using two-dimensional gel electrophoresis reveals distinct differential expression of storage, anti-nutritional, and allergenic proteins. Plant Sci 2008;175:321-9. [CrossRef]
35. Yousuf PY, Ahmad A, Ganie AH, Iqbal M. Salt stress-induced modulations in the shoot proteome of Brassica juncea genotypes. Environ Sci Polut Res 2016;23:2391-401. [CrossRef]
36. Wang L, Liu X, Liang M, Tan F, Liang W. Proteomic analysis of salt-responsive proteins in the leaves of mangrove Kandelia candel during short-term stress. PLoS One 2014;9:1-15. [CrossRef]
37. Page D, Gouble B, Valot B, Bouchet JP, Callot C, Kretzschmar A. Protective proteins are differentially expressed in tomato genotypes differing for their tolerance to low-temperature storage. Planta 2010;232:483-500. [CrossRef]
38. Manaa A, Ben Ahmed H, Valot B, Bouchet JP, Aschi-Smiti S, Causse M. Salt and genotype impact on plant physiology and root proteome variations in tomato. J Exp Bot 2011;62:2797-813. [CrossRef]
39. Gomathi R, Vasantha S, Shiyamala S, Rakkiyappan P. Differential accumulation of salt induced proteins in contrasting sugarcane genotypes. EJBS 2013;6:7-11.
40. Majoul T, Bancel E, Triboi E, Ben Hamida J, Branlard G. Proteomic analysis of the effect of heat stress on hexaploid wheat grain: Characterization of heat-responsive proteins from non-prolamins fraction. Proteomics 2004;4:505-13. [CrossRef]
41. Mann M, Hendrickson RC, Pandey A. Analysis of proteins and proteomes by mass spectrometry. Annu Rev Biochem 2001;70:437-73. [CrossRef]
42. Lee DG, Park KW, An JY, Sohn YG, Ha JK, Kim HY, et al. Proteomics analysis of salt-induced leaf proteins in two rice germplasms with different salt sensitivity. Can J Plant Sci 2011;91:337-49. [CrossRef]
43. Komatsu S, Tanaka N. Rice proteome analysis: A step toward functional analysis of the rice genome. Proteomics 2004;4:938-49. [CrossRef]
44. Sekhwal MK, Swami AK, Sarin R, Sharma V. Identification of salt treated proteins in sorghum using gene ontology linkage. Physiol Mol Biol Plants 2012;18:209-16. [CrossRef]
45. Wang J, Meng Y, Li B, Ma X, Lai Y, Si E, et al. Physiological and proteomic analyses of salt stress response in the halophyte Halogeton glomeratus. Plant Cell Environ 2015;38:655-69. [CrossRef]
46. Noctor G, Queval G, Gakiere B. NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J Exp Bot 2006;57:1603-20. [CrossRef]
47. Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: The PEST hypothesis. Science 1986;234:364-8. [CrossRef]
48. Kirchhoff H, Hall C, Wood M, Herbstova M, Tsabari O, Nevo R. Dynamic control of protein diffusion within the granal thylakoid lumen. Proc Natl Acad Sci USA 2011;108:20248-53. [CrossRef]
49. Krishnasamy L, Selvam MM, Jayanthi K. Novel in silico approach of anticancer activity by inhibiting hemopexin proteins with Indigofera aspalathoides plant constituents at active site. Asian J Pharm Clin Res 2015;8:159-64.
50. Vidhya VG, Upgade A, Bhaskar A, Dipanjana D. In silico characterization of Bovine (Bos taurus) antiapoptotic proteins. J Proteins Proteom 2012;3:187-96.
51. Neelamathi E, Vasumathi E, Bagyalakshmi S, Kannan R. In silico prediction of structure and functional aspects of a hypothetical protein of Neurospora crassa. J Cell Tissue Res 2009;9:1889-94.
52. Ulmschneider MB, Sansom MS. Amino acid distributions in integral membrane protein structures. Biochim Biophys Acta Biomembr 2001;1512:1-14. [CrossRef]
53. Bansal H, Narang D, Jabalia N. Computational characterization of antifreeze proteins of Typhula ishikariensis gray snow mould. J Proteins Proteom 2014;5:169-76.
54. Colovos C, Yeates TO. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci 1993;2:1511-9. [CrossRef]
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Year
Month