1. INTRODUCTION
Groupers are marine and estuarine fishes representing about 163 species in 16 genera distributed worldwide in the tropical and sub-tropical waters. The sub-family Epinephelinae comprises the large commercially important groupers. Around 54 grouper species of the sub-family Epinephelinae and 38 species of the genus Epinephelus have so far been recorded from the Indian waters [1]. Groupers inhabit a diverse range of habitats, mainly coral reefs, rocky areas, seagrass beds, and estuaries [2]. It also exhibits a wide variety of reproductive and growth strategies [3,4]. It was estimated that grouper fish landings in India have been increasing steadily over the years and the output in 2018 was 51,433 tons [5]. The identification characteristics to distinguish the grouper species are usually based on the color pattern and morphological features [6]. However, taxonomic confusion occurred in identifying individual species of the genus Epinephelus due to subtle variations in morphological characteristics [7] and specialties, as well as with a vast number of species [8]. The misidentification of species due to species complexity and synonymies, in addition to the doubts on some generic placements, makes fisheries management and conservation difficult [9,10]. Due to overfishing, some species of groupers are under the threat of extinction. Among 163 species reported worldwide, the International Union for Conservation of Nature (IUCN) Red List of threatened species has categorized 20 species on the verge of extinction and 22 species are considered as nearly threatened [11]. Thus, accurate species identification is important for the analysis of population dynamics, biodiversity assessment, and fishery management.
Epinephelus species are frequently landed in commercial marine catches off Nizampatnam, east coast of India. In spite of their abundance and commercial value, the data on the population characteristics and their conservation status are meager. Furthermore, systematic scientific study has not been conducted so far, including the taxonomic identification of Epinephelus species occurring at Nizampatnam coast. Earlier studies have recorded the prevalence of Epinephelus chlorostigma and Epinephelus bleekeri along the Visakhapatnam coast of India [12]. However, there were reports on the misidentification of Epinephelus areolatus as E. chlorostigma or E. bleekeri because they look similar with overlapping distributions [6,13,14]. In addition to the color pattern and morphological characteristics, the species-specific characteristics, like pyloric caeca [6,15], scales, and otoliths [16–18], and vertebrae [19–21], are proved to be more useful for the identification of species. Furthermore, the advanced molecular technique, such as DNA barcoding, helps in resolving the ambiguity at species-level identification [22–27]. Hence, this study has been carried to identify and validate E. areolatus along Nizampatnam coast, Andhra Pradesh, India, using morpho-meristic characteristics, including pyloric caeca, vertebrae, scales, otoliths, and DNA barcoding. The genetic divergence among the three species of Epinephelus was evaluated using K2P model and the phylogenetic relationships were analyzed based on COI sequence data.
2. MATERIALS AND METHODS
2.1. Sample Collection
Thirty-eight specimens of E. areolatus (n = 38) were collected from the Nizampatnam fish landing center (15° 52′ 58″ N and 80° 38′ 18″ E), Guntur district, Andhra Pradesh, India (Fig. 1), during December 2016 to September 2018, except during the fishing ban period (from April to June). Specimens were observed in fresh condition for color pattern and were photographed (Fig. 2). About 2 g of muscle tissue was collected and preserved in 95% ethanol for molecular analysis and the rest of the specimens were preserved in 5% formalin for morphological study.
2.2. Morphological Analysis
Morpho-meristic data of the species were recorded following the standard methods [6 and 28]. Measurements for morphometric characteristics were taken with a digital vernier caliper to the nearest 1 mm. Vertebrae were counted by radiograph imaging through a digital X-ray. Pyloric caeca were counted at their free ends and fresh coloration was noted. Scales were obtained from eight regions of the fish body (Fig. 3) according to the suggested methods [29,30]. Fine forceps were used to remove scales from the left side of the fish and then cleared in a solution of 1% potassium hydroxide. Sagittal otoliths were extracted by using “Up through the gills method” [31]. The right and left otoliths were separately considered. Digital images and dimensions of scales and otoliths were taken using a Nikon stereomicroscope (SMZ18) with an integrated DS-L3 DS camera. Voucher specimens were documented in the museum of the Department of Zoology and Aquaculture, Acharya Nagarjuna University, Guntur, India (Voucher No. DNAB-5/MUS/NOV/2018).
 | Figure 1: Sampling station at Nizampatnam fishing harbor. [Click here to view] |
2.3. DNA Extraction and Polymerase Chain Reaction (PCR) Amplification
Genomic DNA was extracted from the muscle tissue using DNeasy Blood and Tissue kit (www.qiagen.com). A partial mitochondrial cytochrome oxidase I (COI) gene was amplified by PCR by using a set of published universal primers [27], i.e., FishF1 (5′-TCA ACC AAC CAC AAA GAC ATT GGC AC-3′) and FishR1 (5′-TAG ACT TCT GGG TGG CCA AAG AAT CA-3′) in Eppendorf Master Cycler (Nexus gradient, 230 V) (Eppendorf, Hamburg, Germany). A total of 20 μl reaction mixture was prepared with 5 μl of PCR Smart Master Mix 2X (www.thermofisher.com), 1 μl of forward primer (10 nm/μl), 1 μl of reverse primer (10 nm/μl), approximately 1 μl of DNA template (0.5–1 μg), and 12 μl dH2O. The thermal profile of the PCR reaction was as follows: an initial denaturation at 94°C for 3 minutes, denaturation at 94°C for 45 seconds, annealing temperature of 50°C for 45 seconds, and elongation temperature of 72°C for 1 minute for 30 cycles, and concluded with a final elongation step at 72°C for 7 minutes, followed by a hold at 4°C. The amplicons were verified on 1.5% agarose gel stained with ethidium bromide (EtBr) (10 mg/ml). Amplicons were sent to Macrogen Inc., Seoul, South Korea (www.macrogen.com), for purification and single direction sequencing following the BigDye terminator method [32].
 | Figure 2: Epinephelus areolatus. [Click here to view] |
 | Figure 3: Image showing eight regions (a–h) from where scales were collected. [Click here to view] |
2.4. Sequence Analysis
The COI partial gene sequence obtained for E. areolatus was subjected to nucleotide Basic Local Alignment Search Tool (BLAST) search to know the sequence identity and was further verified for the presence of internal stop codons by using the translate tool in ExPASy ProtParam tool and submitted to NCBI-GenBank to obtain an accession number. The sequence data of E. areolatus were compared with the morphologically similar E. chlorostigma and E. bleekeri by complementing with sequences acquired from the public domain database, NCBI-GenBank (Table 1). The submitted and retrieved sequences were subjected to alignment using ClustalW analysis tool [33] for comparison. Inter- and intra-species genetic divergences in various hierarchical levels were analyzed using Kimura 2 Parameter method [34], with 1,000 bootstrap replicates. The pairwise deletion option was selected to treat the gaps. The neighbor-joining (NJ) tree was generated among the sequences with a suitable out group (Cephalopholis sonnerati, JQ431575) by K2P method [35], using 5,000 pseudo replicates [36].
 | Table 1: List of species used in phylogenetic analysis with NCBI-GenBank accession numbers. [Click here to view] |
3. RESULTS
The following characteristics were noted: body was moderately elongated; skin was thick and leathery; inter orbital space convex; caniniform teeth in both jaws; preopercle was finely serrated with small spines at its lower angle; scales were ctenoid; pectoral fins were longer than the pelvic and anal fins; slightly convex caudal fin in juveniles, slightly emarginated or truncated in adults; depth of the body was less than head length (HL) and contained 2.5–2.8 times standard length (SL) (Table 2); HL contained 2.8–3.0 times in SL; HL was almost similar to the prepelvic distance; pectoral-fin length contained 1.5–1.6 times in HL; pelvic-fin length contained 2.1–2.4 times in HL. The body, head, and fins had numerous yellowish brown or yellow spots; the largest spots were about the size of the pupil; the dorsal fin was gray with small brown spots; soft dorsal distal edge with a white margin; pectoral fin was paler with dark brown spots; pelvic fin was light with dark brown spots; caudal gray with dark brown spots, rear edges with a white margin; anal base was light gray with no spots.
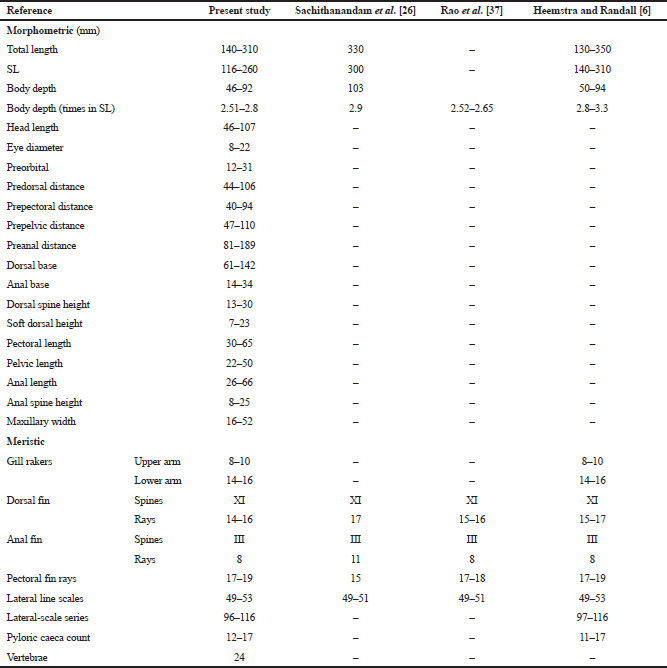 | Table 2: Morphometric and meristic characteristics of E. areolatus (n = 38). [Click here to view] |
The pyloric caecum was present as a whorl in the junction between the stomach and duodenum, mostly on the ventral side, with the flesh colored 12–17 caeca. Vertebrae are 24 (abdominal-10 and caudal-14) (Fig. 4); first two abdominal vertebrae with supraneural spines are directed upward. Scales vary in size in different regions of the body (Fig. 3); the order of scale size is e>h>d>c>g>f>b>a (Table 3 and Fig. 5); 4–8 radii; longer radii are present in scales from regions f to h. Otoliths are slightly oval; right and left otoliths varied in shape and size (Fig. 6 and Table 3). The morphometric and meristic characteristics of the present specimen along with those of earlier works [6,26,37] on this species are shown in Table 2.
 | Figure 4: Radiograph of E. areolatus showing vertebrae. [Click here to view] |
 | Table 3: Scale and otolith dimensions of E. areolatus. [Click here to view] |
 | Figure 5: Scales from different body regions of E. areolatus (a–h) (Scale: a: 500 μm, b–h: 1,000 μm). [Click here to view] |
 | Figure 6: Otolith images of E. areolatus (a-Right; b-Left) (Scale: 1,000 μm). [Click here to view] |
 | Table 4: Intra-species divergence of Epinephelus species using COI gene based on K2P model. [Click here to view] |
 | Table 5: Pairwise inter-genetic distances of Epinephelus species using COI gene based on K2P model. [Click here to view] |
An overall 677 base pair (bp) sequence of mitochondrial COI for E. areolatus was obtained after alignment. The sequence was deposited in NCBI-GenBank with the accession number MK184954. However, the sequences for E. chlorostigma and E. bleekeri were retrieved from the NCBI-GenBank. The multiple sequence alignment led to 649 bp per taxon after eliminating the primer sequence. Sequences were without any ambiguities. No indels (insertions and deletions) and internal stop codons were noted when they were verified in the translate tool using ExPASy ProtParam tool. The pairwise inter- and intra-specific genetic distances among the three species of Epinephelus based on K2P method are presented in Tables 4 and 5. The NJ tree revealed distinct clades for three species with high bootstrap values (Fig. 7).
4. DISCUSSION
Identification of Epinephelus species appears to be challenging because of their closely related morphological characteristics [38–40]. The species E. areolatus has usually been confused with E. chlorostigma and E. bleekeri, and these three species are considered as a single species in the study area. He et al. [13], Heemstra and Randall [6], and Sanaye [14] also reported that these three species are often misidentified as single species due to homogeneity in their morphological characteristics. The morpho-meristic analysis showed some valid indices for species identification. E. areolatus can be distinguished by the rounded margin of the anal fin vs. angular or pointed in E. chlorostigma and E. bleekeri; 14–16 dorsal fin rays vs. 16–18 in E. chlorostigma and E. bleekeri [6]; and 12–17 pyloric caeca vs. 26–52 in E. chlorostigma [6] and 26–36 in E. bleekeri [41]. Moreover, the color configuration of these three species has also been considered for species identification. E. areolatus can be identified by its whitish to gray color with numerous yellowish brown or yellow spots, and caudal fin with a white margin. E. bleekeri has a grayish brown color with numerous orange to orangish red spots, caudal fin with two different colors, upper side yellow, and lower side dusky, whereas E. chlorostigma has a pale green gray color body with small circular yellow or yellowish brown to dark brown spots, caudal fin with a white rear margin [6].
Identification of groupers based on morphological characteristics may be inconclusive [7,22,24,26,41] and even a single systematic external morphological characteristic has not yet been found to differentiate these commercially important groupers [6]. Hence, in this study, to overcome the taxonomical confusion raised by the overlapping characteristics of closely allied Epinephelus species, an attempt has been made by using DNA barcoding as a promising molecular approach to discriminate the species. The barcode established for E. areolatus (MK184954) in this study is the first sequence submitted to the NCBI-GenBank from Nizampatnam coastal waters. The gene sequence for E. areolatus is lacking Indels (insertions and deletions) signifying that the sequence does not comprise nuclear mitochondrial DNA (NUMT). The sequence of E. areolatus matches 98.79% with the sequences of E. areolatus (KU499782 and KU499599) accessible in the NCBI-GenBank. The mean intraspecific genetic divergence analysis revealed higher values in E. chlorostigma (0.032 ± 0.004), followed by E. areolatus (0.012 ± 0.003) and E. bleekeri (0.001 ± 0.001) (Table 4). The mean interspecific genetic distance showed high similarity with E. bleekeri (0.109 ± 0.012) and low similarity with E. chlorostigma (0.079 ± 0.009) (Table 5). The phylogenetic analysis from NJ tree clearly shows the genetic divergence of the three species with high bootstrap values (Fig. 7). Hence, it is evident from this study that E. areolatus, E. bleekeri, and E. chlorostigma are distinctly valid species and are clearly separable from other congeners.
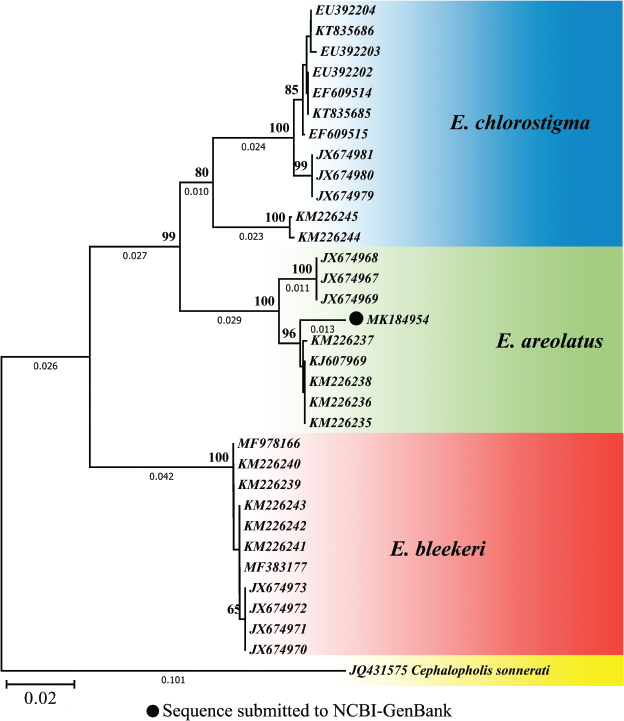 | Figure 7: Neighbor-joining tree of COI gene sequences for Epinephelus species. [Click here to view] |
5. CONCLUSION
Perfect taxonomic identification of species is crucial for proper management of any fishery resource. In this study, the morphological data of E. areolatus were cataloged to differentiate with other congeneric species, more precisely with reference to the shape of the anal fin, dorsal fin rays, and pyloric caeca. The COI gene sequence and phylogenetic analysis revealed genetic divergence and relatedness of the three species. Hence, both morphological data and gene sequencing are crucial for the validation of species and will continue to improve our understanding in the identification of grouper fishes. The occurrence and genetic diversity of Epinephelus in Indian waters need further investigation for more insights in view of their nutritional and commercial importance.
ACKNOWLEDGMENTS
The authors are thankful to the authorities of Acharya Nagarjuna University for providing the basic facilities to execute this work in the Department of Zoology and Aquaculture. We are thankful to Dr. Ritesh Ranjan, Mr. P. Shiva, and Mr. B. Vamsi, CMFRI, Visakhapatnam, India, for providing the microscope facility.
CONFLICT OF INTEREST
Authors declared that there is no conflicts of interest.
REFERENCES
1. Froese R, Pauly D. [Internet]. FishBase. 2019. Available via www.fishbase.org (Accessed 18 December 2019).
2. Rao DV. Checklist of fishes of Andaman and Nicobar Islands, Bay of Bengal. Environ Ecol 2009;27(1A):334–53.
3. Morris AV, Roberts CM, Hawkins JP. The threatened status of groupers (Epinephelinae). Biodivers Conserv 2000;9(7):919–42. CrossRef
4. Shapiro DY. Reproduction in groupers. In: Polovina JJ and Ralston S (eds.). Tropical snappers and groupers: biology and fisheries management. Westview Press, London, UK, pp 295–327, 1987.
5. FRAD, CMFRI. Marine fish landings in India 2018. Technical Report, CMFRI, Kochi, India, 2019.
6. Heemstra PC, Randall JE. Groupers of the world. In: FAO Species Catalogue. Groupers of the world (Family Serranidae, Subfamily Epi- Authors nephelinae). An annotated and illustrated catalogue of the grouper, rockcod, hind, coral grouper and lyretail species known to date. FAO Fisheries Synopsis, Rome, Italy, p 382, 1993.
7. Darwin CH, Padmavathi P, Srinu G. DNA divergence and genetic relatedness of Epinephelus species (Perciformes:Serranidae) of Indian waters inferred from COI sequence data. In: Vijaya N, (ed.). Recent trends in applied biology. Adikavi Nannaya University, Rajamahendravaram, India, pp 3–16, 2018.
8. Craig MT, Pondella II DJ, Franck JP, Hafner JC. On the status of the serranid fish genus Epinephelus: evidence for paraphyly based upon 16S rDNA sequence. Mol Phylogenet Evol 2001;19(1):121–30. CrossRef
9. Craig MT, Hastings PA. A molecular phylogeny of the groupers of the subfamily Epinephelinae (Serranidae) with a revised classification of the Epinephelini. Ichthyological Res 2007;54(1):1–7; doi:10.1007/s10228-006-0367-x
10. Schoelinck C, Hinsinger DD, Dettai A, Cruaud C, Justine JL. A phylogenetic re-analysis of groupers with applications for ciguatera fish poisoning. PLoS One 2014;9(8):e98198; doi:10.1371/journal. pone.0098198
11. Sadovy de Mitcheson Y, Craig MT, Bertoncini AA, Carpenter KE, Cheung WW, Choat JH, et al. Fishing groupers towards extinction: a global assessment of threats and extinction risks in a billion dollar fishery. Fish Fish 2013;14(2):119–36; doi: 10.1111/j.1467-2979.2011.00455.x
12. Kandula S, Shrikanya KV, Iswarya Deepti VA. Species diversity and some aspects of reproductive biology and life history of groupers (Pisces: Serranidae: Epinephelinae) off the central eastern coast of India. Mar Biol Res 2015;11(1):18–33; doi:10.1080/17451000.2014.949271
13. He B, Lai T, Peng Z, Wang X, Pan L. Complete mitogenome of the areolate grouper Epinephelus areolatus (Serranidae, Epinephelinae). Mitochondrial DNA 2013;24(5):498–500; doi:10.3109/19401736.2013.770503
14. Sanaye SV [Internet]. India biodiversity portal. 2018. Available via https://indiabiodiversity.org/ (Accessed 18 December 2019).
15. Roy TS, Gopalakrishnan A. Identification of groupers based on pyloric caeca differentiation. J Fish Biol 2011;79(5):1334–9; doi:10.1111/j.1095-8649.2011.03089.x
16. Chervinski J. Using scales for identification of four Mugilidae species. Aquac 1984;38(1):79–81; doi:10.1016/0044-8486(84)90139-X CrossRef
17. Ibáñez AL, O’Higgins P. Identifying fish scales: the influence of allometry on scale shape and classification. Fish Res 201;109(1):54–60; doi:10.1016/j.fishres.2011.01.016 CrossRef
18. Popper AN, Ramcharitar J, Campana SE. Why otoliths? Insights from inner ear physiology and fisheries biology. Mar Freshw Res 2005;56(5):497–504. CrossRef
19. Jawad LA. A comparative morphological investigation of otoliths of six parrotfish species (Scaridae) from the Solomon Islands. J Fish Biol 2018;93(6):1046–58; doi:10.1111/jfb.13787
20. Jawad LA, Al-Hassani L. Morphological study of the vertebral column of the ponyfish Leiognathus equulus (Family: Leiognathidae) collected from the sea of Oman. Int J Mar Sci 2014;4(33):1-6; doi: 10.5376/ ijms.2014.04.0033.
21. Kubo Y, Asano H. Relative growth pattern and hard tissue of vertebral centra by microradiography of bluefin tuna. Nippon Suisan Gakkai-Shi 1989;56(7):1021–7. CrossRef
22. Basheer VS, Vineesh N, Bineesh KK, Kumar RG, Mohitha C, Venu S, et al. Mitochondrial signatures for identification of grouper species from Indian waters. Mitochondrial DNA Part A 2017;28(4):451–7; doi:10.3109/19401736.2015.1137899
23. Hebert PD, Cywinska A, Ball SL, Dewaard JR. Biological identifications through DNA barcodes. Proc R Soc Lond B Biol Sci 2003;270(1512):313–21. CrossRef
24. Iswarya Deepti V, Kandula S, Khedkar GD. DNA barcoding of five species of groupers (Pisces: Serranidae) off Visakhapatnam, central eastern coast of India. Mitochondrial DNA Part A 2018;29(5):659–63; doi:10.1080/24701394.2017.1339188
25. Noikotr K, Chaveerach A, Pinthong K, Tanomtong A, Sudmoon R, Tanee T. RAPD and barcode analyses of groupers of the genus Epinephelus. Genet Mol Res 2013;12(4):5721–32; doi:10.4238/2013
26. Sachithanandam V, Mohan PM, Muruganandam N, Chaaithanya IK, Dhivya P, Baskaran R. DNA barcoding, phylogenetic study of Epinephelus spp. from Andaman coastal region, India. Indian J Geo Mar Sci 2012;41(3):203–11.
27. Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PD. DNA barcoding Australia's fish species. Philos Trans R Soc B 2005;360(1462):1847–57. CrossRef
28. de Mitcheson YS, Heemstra PC. Groupers of the world: a field and market guide. In: Craig MT, (ed.). Grahamstown (South Africa): CRC Press, Boca Raton, FL, 2011.
29. Jawad LA. Comparative morphology of scales of four teleost fishes from Sudan and Yemen. J Nat Hist 2005;39(28):2643–60; doi:10.1080/00222930500102801
30. Jawad LA, Al-Jufaili SM. Scale morphology of greater lizardfish Saurida tumbil (Bloch, 1795) (Pisces: Synodontidae). J Fish Biol 2007;70(4):1185–212; doi:10.1111/j.1095-8649.2007.01385.x.
31. Secor DH, Dean JM, Laban EH. Manual for otolith removal and preparation for microstructural examination. Copeia 1992;2:85 CrossRef
32. Rosenblum BB, Lee LG, Spurgeon SL, Khan SH, Menchen SM, Heiner CR, et al. New dye-labeled terminators for improved DNA sequencing patterns. Nucleic Acids Res 1997;25(22):4500–4; doi:10.1093/nar/25.22.4500 CrossRef
33. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997;25(24):4876–82; doi:10.1093/nar/25.24.4876 CrossRef
34. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16(2):111–20. CrossRef
35. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4(4):406–25.
36. Phylip JF. [Internet]. Phylogeny inference package version 3.6. 2004. Available via http://www.evolution.gs.washington.edu/phylip.html (Accessed 18 December 2019).
37. Rao DV, Kamala D, Ranjan PT. An account of ichthyofauna of Andaman & Nicobar Islands, Bay of Bengal, records of the zoological survey of India. Occ Paper No 2000;178:1–434.
38. Mekkawy IA, Mohammad AS. Morphometrics and meristics of the three epinepheline species: Cephalopholis argus (bloch and schneider, 1801), Cephalopholis miniate (Forsskal, 1775) and Variola louti (Forsskal, 1775) from the Red Sea, Egypt. J Biol Sci 2011;11(1):10–21. CrossRef
39. Mekkawy IA, Saber SA, Shehata SM, Osman AG. Morphometrics and meristics of four fish species of genus Epinephelus (Family Seranidae) from the Red Sea, Egypt. Bull Fac Sci Assiut Univ 2002;31:21–41.
40. Sujatha K. Groupers off Visakhapatnam, north east coast of India. J Mar Biol Assoc India 2004;46(1):87–92.
41. Deepti VA, Shrikanya KV, Sujatha K. Taxonomic studies and phylogenetic relationship of seven spotted groupers species of genus Epinephelus (Pisces: Serranidae) off Visakhapatnam, middle east coast of India. Indian J Geo-Marine Sci 2014;43(12):2254–68.