1. INTRODUCTION
Proteases lead the worldwide industrial enzyme market and account for nearly 60% of the total worldwide enzyme sales [1-4]. Proteases can be classified according to their active pH range into neutral, acidic, and alkaline pH. Each of these groups is also divided into six subclasses based on the type of functional groups. Alkaline proteases such as Subtilisin, Carlsberg, Alcalase, Esperase, and Savinase are produced by bacteria, in particular, Bacillus sp. are commercial proteases [5-7]. Several fungi including Aspergillus sp. are also considered to be a good source of protease production [8]. There is very less information available on naturally occurring extracellular proteases produced by actinomycetes. For industrial production, only the microbes that produce a large amount of extracellular protease can be exploited. Proteases produced from actinomycetes have unique characteristics [9-11] and should be investigated so as to meet the requirements of different industrial processes [12]. Medium formulation and optimization increase the production of protease [13-14]. The purification process was found to increase the specific activity of enzymes and to make them more acceptable for industrial applications [15,16].
2. MATERIALS AND METHODS
2.1. Isolation of Streptomyces sp. LCJ12A
The source of Streptomyces sp. LCJ12A was the soil of Pichavaram Mangrove Forest, Chidambaram, Tamil Nadu, India, located at the latitude of 11.42714 and longitude position of 79.78326.
One gram of soil sample collected from Pichavaram Mangrove Forest was serially diluted and 100 μl of 10−4 dilution was plated on International Streptomyces Project-2 medium. The plates were then incubated at 30°C for 12 days. After incubation, the pure cultures were maintained at 4°C.
2.2. Protease Production under Submerged Fermentation
Protease production broth (PPB) medium was used as a production medium for the production of protease. Spore suspension (1 mL) of the isolate strain was added to a 250 mL flask containing 50 mL of PPB containing (g/L) peptone – 10, sucrose – 10, K2HPO4 – 0.5, CaCl2 – 0.5, NaCl – 0.5, and MgSO4.7H2O – 0.5. The mixture was incubated under shaking conditions at 30°C and 120 rpm. Once in 2 days, the cultures were harvested and the culture medium was centrifuged to remove medium debris and mycelia. Quantitative enzyme assay was used to determine the protease activity of culture supernatants (Ramesh et al., 2009).
2.3. Protease Purification
The culture filtrate from an 8-day-old culture was harvested and primarily filtered through a muslin cloth to eliminate the mycelial debris. The filtrate was centrifuged at 10,000 rpm for 15 min at 4°C and again it was filtered through a Whatman No. 1 filter paper.
2.4. Ammonium Sulfate Precipitation
The filtered supernatant of Streptomyces sp. LCJ12A was subjected to precipitation using ammonium sulfate at 70% (w/v) saturation at 4°C overnight. Centrifugation (10,000 rpm for 15 min) was used to collect the precipitate and again resuspended in sodium acetate buffer (100 mM; pH 5.0). This was dialyzed against the same buffer overnight at 4°C. After that, the dialyzed filtrate was loaded onto the diethylaminoethyl (DEAE) cellulose ion-exchange column.
2.5. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
Clean glass plates were kept in vertical position by placing them in a gel casting stand. The separating gel mixture was prepared and poured into the glass plates uniformly without any leakage. The stacking gel was prepared and poured over it and the comb was inserted gently on the stacking gel after the polymerization of the separating gel. To allow the gel to polymerize completely, the setup was kept for few minutes. The comb was then carefully removed and wells formed in the gel were rinsed with distilled water completely. 50 μl of isolated protein samples was mixed with 50 μl of sample solubilizing buffer (1:1) and the mixture was boiled at 95°C in water bath for 5–10 min. The protein samples along with standard protein marker were then loaded into the respective wells. To allow the sample to run completely into the gel, it was electrophoresed at 50–100 v for 3–4 h. Coomassie brilliant blue (CBB) R250 was used as a staining solution and the gel was placed in CBB for 1 h after electrophoresis. After staining, the gel was washed thoroughly with distilled water. To destain the stained gel, the stained gel was placed in destaining solution for overnight and washed with distilled water [17]. The proteins fractionated into bands were observed and the molecular weight of the protein was also determined using standard marker.
2.6. Ion Exchange Column Chromatography
The dialyzed filtrate was loaded onto a DEAE cellulose ion exchange column pre-equilibrated with glycine-NaOH buffer (100 mM; pH 8.6) for Streptomyces sp. LCJ12A which was washed with the same respective buffers to remove unbound proteins. The bound proteins were then eluted with a linear gradient of NaCl (0–1.0 M) in the respective buffers at a flow rate of 1 mL/min and the fractions were collected and assayed for protease activity. The protease rich fractions were pooled, concentrated, and dialyzed against the respective buffers. The total specific activity, yield, and purification fold were calculated according to the standard method [18].
2.7. Molecular Mass Determination of Protease
SDS-PAGE was carried out in accordance with the standard protocol using 10% w/v resolving gel and 5% w/v stacking gel. Separation of the proteins was carried out at 20 mA. After electrophoresis, CBB R250 was used to stain the protein bands. The molecular mass of the partially purified protein was determined by comparison against the relative mobility of a broad range of standard protein markers (Genie, India), namely, phosphorylase B (97.4 kDa), ovalbumin (43 kDa), bovine serum albumin (BSA-66 kDa), soybean trypsin inhibitor (20.1 kDa), carbonic anhydrase (29 kDa), and lysozyme (14.3 kDa).
2.8. Influence of pH on Protease Activity
The optimum pH of the partially purified protease, culture filtrate, and ammonium sulfate precipitate was determined over a range of pH (3–11.5). The partially purified enzyme (10 μg) was incubated for 5 min in different buffer systems such as glycine-HCl buffer (100 mM; pH 2.2–3.4), sodium acetate buffer (100 mM; pH 3.8–5.4), phosphate buffer (100 mM; pH 5.8–7.8), and glycine-NaOH buffer (100 mM; pH 8.2–11.5). The substrate was then added to the reaction mixture and assayed for protease activity. Similarly, culture filtrate and ammonium sulfate precipitate were also assayed at different pH.
2.9. Influence of Temperature on Protease Activity and Thermal Stability
The influence of temperature on the protease activity and its stability was determined by dissolving the partially purified protease (10 μg) in glycine NaOH (100 mM; pH 8.6) for Streptomyces sp. LCJ12A which was incubated separately at temperatures ranging from 25°C to 50°C for 5 min and the protease activity was determined. Culture filtrate and ammonium sulfate precipitate were also assayed for protease activity. The thermal stability of the protease enzyme was investigated by measuring the protease activity and it was done after the pre-incubation of the partially purified protease (10 μg) for 1 h at temperatures ranging from 25 to 50°C.
2.10. Substrate Specificity
Substrate specificity of the partially purified protease enzyme, culture filtrate, and ammonium sulfate precipitate was determined using various substrates such as BSA, casein, and gelatin at a concentration of 1 mM. The 10 μg of partially purified protease, culture filtrate, and ammonium sulfate precipitate were incubated with the above substrates and their oxidation was measured in their respective absorption maxima. The protease activity was calculated by means of the respective extinction coefficient values.
2.11. Kinetic Constants of Partially Purified Protease
The maximum velocity (Vmax) and Michaelis–Menten constant (Km) are the most important parameters for most of the enzyme-catalyzed reactions. Studies on enzyme kinetics were conducted using casein, BSA, and gelatin in 100 mM phosphate buffer (pH 5.0). The Km value of enzyme determines the affinity of the enzyme for its substrate [19]. Microsoft Excel was used to calculate linear regression for estimating Vmax and Km. Reciprocal plot of velocity of the reaction and substrate concentrations were plotted using Lineweaver–Burk plot. The maximum velocity and Michaelis constant were calculated according to the method described by Nelson and Cox, 2004. The formula used is mentioned below:
- Maximum Velocity (Vmax): The reciprocal of the intercept on the positive Y-axis is the Vmax (mM min−1)
- Michaelis constant (Km): The reciprocal of the intercept on the negative X-axis is the Km (mM).
3. RESULTS
The molecular mass of the partially purified protease from Streptomyces sp. LCJ12A was determined by SDS-PAGE. The partially purified protease appeared as a single band on the SDS-PAGE stained with CBB R250 and its molecular weight was approximately 20.1 kDa in comparison with the standard molecular weight markers. The results indicate the monomeric nature of the partially purified protease from Streptomyces sp. LCJ12A [Figure 1].
3.1. Influence of pH on Protease Activity
The partially purified protease, culture filtrate, and ammonium sulfate precipitate from Streptomyces sp. LCJ12A were evaluated under different pH conditions ranging from 3 to 11.5. The partially purified protease from Streptomyces sp. LCJ12A demonstrated 100% of relative activity at pH 10 when casein was used as the substrate [Figure 2]. Culture filtrate with 90% relative activity and ammonium sulfate precipitate with 93% relative activity were observed at pH 10. However, considerable relative activity was observed between pH 6.5 and 8.0 with the relative activity ranging from 65 to 90% for partially purified protease. At pH range of 5.0–3.5, less relative activity was observed ranging from 40 to 20%. In Streptomyces sp. LCJ12A, the optimum pH value was recorded as pH 10. Further increase or decrease in pH resulted in a lesser relative activity.
3.2. Effect of Different Temperature and Temperature Stability on Protease Activity
The effect of various temperatures on the partially purified protease, culture filtrate, and ammonium sulfate precipitate from Streptomyces sp. LCJ12A was determined at temperature ranging from 25 to 50°C. The optimum temperature of partially purified protease was found to be 35°C with a maximum relative activity of 100% [Figure 3] and it was furthermore significant.
.png) | Figure 1: Molecular mass determination of protease from Streptomyces sp. LCJ12A on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. [Click here to view] |
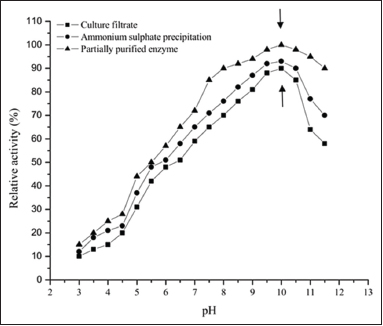 | Figure 2: Relative activity of protease enzyme at different pH from Streptomyces sp. LCJ12A. [Click here to view] |
 | Figure 3: Relative activity of protease enzyme at different temperature from Streptomyces sp. LCJ12A. [Click here to view] |
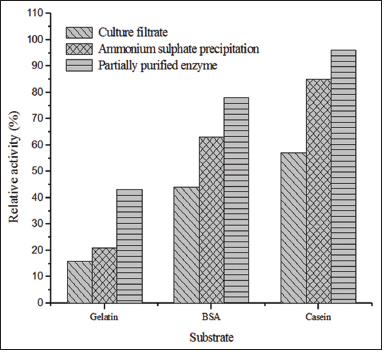 | Figure 4: Substrate oxidizing activity of protease enzyme from Streptomyces sp. LCJ12A. [Click here to view] |
Temperature stability of the partially purified protease, culture filtrate, and ammonium sulfate precipitate from Streptomyces sp. LCJ12A was evaluated at temperatures ranging between 25°C and 50°C. The protease activity demonstrated a smooth and stable increase as the temperature increased from 30°C to 40°C. The optimum temperature for partially purified protease was found to be at 35°C with an initial activity of 85% at 10 min, and it retained 100% of the relative activity and was stable at the end of 60 min. Considerable amount of relative activity (92%) was also observed at 40°C. The protease activity decreased drastically when the temperature exceeded 45°C. The partially purified protease from Streptomyces sp. LCJ12A retained only 78% of its activity after incubation at 45°C for 20 min. When the temperature was increased, further, the activity of the enzyme decreased rapidly and 60% relative activity was observed at the end of 1 h at 50°C. The relative activity decreased drastically as the enzyme was incubated for a longer time in these temperatures. The results indicate that the protease was not thermostable. The statistical analysis further confirmed that 35°C was significant for partially purified enzyme, culture filtrate, and ammonium sulfate precipitate when compared to other temperatures.
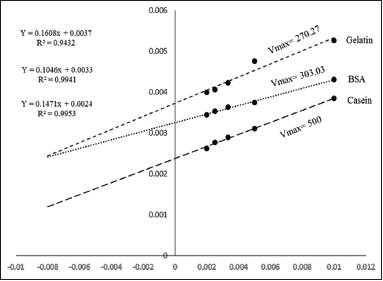 | Figure 5: Lineweaver–Burk plot of partially purified protease from Streptomyces sp. LCJ12A. [Click here to view] |
 | Table 1: Kinetic constant of partially purified protease from Streptomyces sp. LCJ12A [Click here to view] |
3.3. Substrate Specificity
The substrate oxidization activity of the partially purified protease enzyme, culture filtrate, and ammonium sulfate precipitate from Streptomyces sp. LCJ12A toward various substrates such as BSA, casein, and gelatin was characterized. Among the substrates tested, casein was the most efficiently oxidized substrate with maximum relative activity of 96% of partially purified protease. The relative activity of the culture filtrate and ammonium sulfate precipitate was 85% and 57%, respectively [Figure 4]. The partially purified protease from Streptomyces sp. LCJ12A demonstrated substantial relative activity of 78% with BSA and 43% with gelatin. The statistical analysis also confirmed that compared to the other substrates, casein was significant.
3.4. Kinetic Constants of Partially Purified Protease
The kinetic constants of partially purified protease from Streptomyces sp. LCJ12A were determined using casein, BSA, and gelatin. The Km and Vmax values for the partially purified protease were calculated from the Lineweaver–Burk plot and presented in Figure 5. The Km values of Streptomyces sp. LCJ12A were 73.5 mM for casein, 44.54 mM for BSA, and 43.45 mM for gelatin. The Vmax values were 500 mM min−1 for casein, 303.03 mM min−1 for BSA, and 270.27 mM min−1 for gelatin for Streptomyces sp. LCJ12A which are summarized in Table 1.
4. DISCUSSION
The studies related to the actinomycetes from soil and sediments of Pichavaram mangrove region are very less explored. Actinomycetes are good sources for biotechnological applications [20]. Ion-exchange chromatography was used on a DEAE cellulose column. The culture filtrate was subjected to ammonium sulfate precipitation (70%) followed by ion-exchange chromatography in CM-650 TOYOPEARL® column. The appearance of single band in SDS indicated that the purified alkaline protease was a monomeric protein with molecular mass of 20.1 kDa. In general, the molecular mass of alkaline proteases from microbe ranges 15 to 40 kDa. Alkaline proteases have been purified from other microbes and have been characterized and their molecular weight is as follows: Streptomyces albidoflavus (18 kDa) [21], Streptomyces sp. M30 (37.1 kDa) [22], Bacillus circulans MTCC 7942 (43 kDa) [23], Aspergillus parasiticus (36 kDa) [24], and Thermus aquaticus YT-1 (38 kDa) [25]. The molecular mass of alkaline proteases from Bacillus genus has been found to be above 70 kDa [26-28]. Alkaline proteases from Bacillus laterosporus AK1 having molecular weight above 70 kDa were also reported [29-31].
The purification and characterization of a 20.1 kDa alkaline protease from Streptomyces sp. LCJ12A are described in this investigation. Proteolytic activity in this study increased up to 100% for partially purified enzyme at the temperature of 35°C when casein was used as a substrate and the proteolytic activity decreased dramatically when the temperature increased above 40°C, which retained only 78% at 45°C and 60% activity at 50°C, respectively. The proteolytic activity of culture filtrate and ammonium sulfate precipitate was 90% and 96% at 35°C. This was similar to the results reported [32], where about 95% and 70% activity of Bacillus protease were retained at 40°C and 50°C.
The optimal pH for the higher proteolytic activity from Streptomyces sp. LCJ12A was pH 10. Streptomyces sp. LCJ12A showed 100% of proteolytic activity at pH 10 and further increase in pH decreased the proteolytic activity from 100% to 90% for partially purified enzyme. The proteolytic activity of culture filtrate and ammonium sulfate precipitate was 90% and 93% at pH 10. The study from Streptomyces clavuligerus NRRL 3585 and 644 reported that the proteolytic activity of 37% obtained at pH 8.0–8.4 for partially purified enzyme [33]. For the enzyme keratinase, the maximal enzyme activity was 80% at pH 11 at 45°C from Streptomyces sp. [34].
The kinetic constant of partially purified enzyme from Streptomyces sp. LCJ12A showed Vmax of 500 mM min−1 and Km of 73.5 mM for casein substrate when BSA used as substrate, Vmax 303.03 mM min−1 and Km 44.54 mM were recorded. Vmax of 270.27 mM min−1 and Km of 43.45 mM were recorded for gelatin as a substrate. Similar enzyme kinetic study was also done by proteolytic bacteria showed 0.3 and 0.5 mg ml-1 and Vmax 333 and 500 μmole min−1 [35].
5. CONCLUSION
The findings of the current study show that the enzyme had shown broad substrate specificity. The enzyme exhibited high activity at alkaline pH and moderate temperature (below 40°C) inferred that the purified enzyme seems to be potential for industrial applications. The partially purified protease enzyme from Streptomyces sp. LCJ12A acts as a potential thrombolytic agent having fibrinolytic activity, dissolves the coagulated sheep blood, and also has the potential toward blood stain removal from the cotton fabrics and dehairing of goat hide. In the next phase of study, we have also identified the suitable pH and temperature of the protease to improve the efficiency of fibrinolytic activity, blood stain removal, and dehairing. This study suggests that the enzyme protease has the ability to be used under industrial circumstances for various applications. The industrial applications of the protease are in progress and will be reported elsewhere.
6. ACKNOWLEDGMENTS
We are thankful to “Times of India” and Loyola College, Chennai, for providing financial support to carry out this study.
7. REFERENCES
1. Kumar CG, Takagi H. Microbial alkaline proteases: From a bioindustrial viewpoint. Biotechnol Adv 1999;17:561-94. CrossRef
2. Maurer KH. Detergent proteases. Curr Opin Biotech 2004;15:330-4. CrossRef
3. Saeki K, Ozaki T, Kobayashi T, Ito S. Detergent alkaline proteases: Enzymatic properties, genes and crystal structures. J Biosci Bioeng 2007;103:501-8. CrossRef
4. Rani K, Rana R, Datt S. Review on latest overview of protease. Int J Curr Life Sci 2012;2:12-8.
5. Sreedevi B, Abhigna A, Kumari JP. Isolation and characterization of alkalophilic Pseudomonas sp. and optimization of culture conditions for alkaline protease production. Environ Res J 2013;7:30-6.
6. Bessler C, Maurer KH, Merkel M, Siegert P, Weber A, Wieland S. Novel Alkaline Protease from Bacillus gibsonii and Washing and Cleaning agents Containing said Novel Alkaline Protease. Patent Appl WO 2007/131657.
7. Henkel AG, Kgaa CO. Novel Alkaline Protease from Bacillus gibsonii and Washing and Cleaning agents Containing said Novel Alkaline Protease. Patent Appl US 2009/0275493.
8. Gnanadoss JJ, Devi SK. Optimization of nutritional and culture conditions for improved protease production by Aspergillus nidulans and Aspergillus flavus. J Microbiol Biotech Food Sci 2015;4:518-23. CrossRef
9. Ramesh S, Rajesh M, Mathivanan N. Characterization of a thermostable alkaline protease produced by marine Streptomyces fungicidicus MML1614. Bioprocess Biosyst Eng 2009;32:791-800. CrossRef
10. Sanatan PT, Lomate PR, Giri AP, Hivrale VK. Characterization of a chemostable serine alkaline protease from Periplaneta americana. BMC Biochem 2013;14:32-4. CrossRef
11. Sharma P, Bajaj BK. Production and partial characterization of alkali-tolerant xylanase from an alkalophilic Streptomyces sp. CD3. J Sci Ind Res 2005;64:688-97.
12. Cowan DA. Thermophilic proteins: Stability and function in aqueous and organic solvents. Comp Biochem Physiol A Physiol 1997;118:429-38. CrossRef
13. Parthasarathy M, Gnanadoss JJ. Medium formulation and its optimization to enhance protease production by Streptomyces sp. Isolated from mangroves. Biosci Biotech Res Asia 2018;15:719-28. CrossRef
14. Parthasarathy M, Gnanadoss JJ. Medium formulation and optimization for improving protease production by Streptomyces sp. LCJ17A. Int J Life Sci Res 2018;6:277-86.
15. Kumar CG. Purification and characterization of a thermostable alkaline protease from alkalophilic Bacillus pumilus. Lett Appl Microbiol 2002;34:13-7. CrossRef
16. Adinarayana K, Ellaiah P, Prasad D. Purification and partial characterization of thermostable serine alkaline protease from a newly isolated Bacillus subtilis PE-1. AAPS Pharm SciTech 2003;4:440-8. CrossRef
17. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227(5259):680-5. CrossRef
18. Nelson DL, Cox MM. Lehninger Principles of Biochemistry. 4th ed. New York, USA: Freeman WH, Co.; 2004. p. 1100.
19. Ortiz GE, Noseda DG, Mora MC, Recupero MN, Blasco M, Alberto EA. Comparative study of new Aspergillus strains for proteolytic enzymes production by solid state fermentation. Enzyme Res 2016;Article ID: 3016149:11. CrossRef
20. Rifaat HM, Osama HE, Sadia MH, Manal SM. Protease activity of some mesophilic Streptomycetes isolated from Egyptian habitats. J Cult Collect 2006;5:16-24.
21. Bressollier P, Letourneau F, Urdaci M, Verneuil B. Purification and characterization of a keratinolytic serine proteinase from Streptomyces albidoflavus. Appl Environ Microbiol 1999;65:2570-6. CrossRef
22. Xin Y, Sun Z, Chen Q, Wang J, Wang Y, Luogong L, et al. Purification and characterization of a novel extracellular thermostable alkaline protease from Streptomyces sp. M30. J Microbiol Biotechnol 2015;25:1944-53. CrossRef
23. Patil U, Mokashe N, Chaudhari A. Detergent-compatible, organic solvent-tolerant alkaline protease from Bacillus circulans MTCC 7942: Purification and characterization. Prep Bioche Biotechnol 2016;46:56-64. CrossRef
24. Anitha TS, Palanivelu P. Purification and characterization of an extracellular keratinolytic protease from a new isolate of Aspergillus parasiticus. Protein Expr Purif 2013;88:214-20. CrossRef
25. Ol?dzka G, D?browski S, Kur J. High-level expression, secretion, and purification of the thermostable aqualysin I from Thermus aquaticus YT-1 in Pichia pastoris. Protein Expr Purif 2003;29:223-9. CrossRef
26. Jung SC, Paik HR, Kim MS, Baik KS, Lee WY, Seong CN, et al. InhA-like protease secreted by Bacillus sp. S17110 inhabited in Turban shell. J Microbiol 2007;45:402-8.
27. Ghosh A, Chakrabarti K, Chattopadhyay D. Degradation of raw feather by a novel high molecular weight extracellular protease from newly isolated Bacillus cereus DCUW. J Ind Microbiol Biotechnol 2008;35:825-34. CrossRef
28. Kato T, Yamagata Y, Arai T, Ichishima E. Purification of a new extracellular 90-kDa serine proteinase with isoelectric point of 3.9 from Bacillus subtilis (natto) and elucidation of its distinct mode of action. Biosci Biotechnol Biochem 1992;56:1166-8. CrossRef
29. Arulmani M, Aparanjini K, Vasanthi K, Arumugam P, Arivuchelvi M, Kalaichelvan PT. Purification and partial characterization of serine protease from thermostable alkalophilic Bacillus laterosporus AK1. World J Microbiol Biotechnol 2007;23:475-81. CrossRef
30. Vidyasagar M, Prakash S, Litchfield C, Sreeramulu K. Purification and characterization of a thermostable, haloalkaliphilic extracellular serine protease from the extreme halophilic archaeon Halogeometricum borinquense strain TSS101. Archaea 2006;2:51-7. CrossRef
31. Singh L, Ramana KV. Isolation and Characterization of Psychrotrophic Antarctic Bacteria from Blue-green Algal Mats and their Hydrolytic Enzymes. Fourteenth Indian Expedition to Antarctica, Scientific Report, Department of Ocean Development, Technical Publication No 12, 1998. p. 199-206.
32. Pawar R, Zambare V, Barve S, Paratkar G. Application of protease isolated from Bacillus sp. 158 in enzymatic cleansing of contact lenses. Biotechnology 2009;8:276-80. CrossRef
33. Moreira KA, Cavalcanti MT, Duarte HS, Tambourgi EB, Melo EH, Silva VL, et al. Partial characterization of proteases from Streptomyces clavuligerus using an inexpensive medium. Braz J Microbiol 2001;32:215-20. CrossRef
34. Tatineni R, Doddapaneni KK, Potumarthi RC, Vellanki RN, Kandathil MT, Kolli N, et al. Purification and characterization of an alkaline keratinase from Streptomyces sp. Bioresour Technol 2008;99:1596-602. CrossRef
35. Iqbal A, Hakim A, Hossain SM, Rahman MR, Islam K, Azim MF, et al. Partial purification and characterization of serine protease produced through fermentation of organic municipal solid wastes by Serratia marcescens A3 and Pseudomonas putida A2. J Genet Eng Biotechnol 2018;16:29-37. CrossRef