INTRODUCTION
Seafood including fish, prawn, shrimp, lobster, crayfish, crab, and clam has become a favorite nutritious delicacy for mankind [1]. They are a good source of complete protein and act as the building blocks necessary for the production of new proteins. Seafood products are frequently mislabeled and misrepresented food items. Mislabeling is sometimes by human error but mostly by intention. Usually, the superior valued species are substituted by substandard ones, which are causing serious health concern in addition to economic implications and financial risks on consumer [2–5]. According to OCEANA, 33% of seafood is mislabeled, which was revealed through DNA-based investigations [6–8]. The mislabeled seafood may be dangerous due to the presence of unknown toxic/allergenic substances. It may cause species loss if an endangered species is commercialized [9,10]. Under sliced, packed, and marketed stage, most of the prawns, shrimps, and fishes are similar in texture and it is very difficult to differentiate the right species without diagnosing either by body parts or DNA analysis.
There are several brands of frozen seafood available in India that are being exported. The majority of brands do not specify the species used. There is a mixing of species in packages. In such cases, for example, minced meat, swapping can be hidden as the seafood is highly processed. The nomenclature of commercially important seafood is not globally standardized; varied species are recognized by the same vernacular name, and a single species may be labeled with different names in different regions of the same country. Therefore, the Food and Drug Administration (FDA) of US (1988) and the EU (CE, 2065/2001) have stressed the scientific community and commercial industries for using adequate tools to confirm species authenticity, products labeling, and avoiding commercial fraud in seafood products labeling [11–16].
DNA barcoding is applied in biodiversity monitoring (taxonomy, ecology, and conservation) to identify organisms that lack distinctive morphological features due to homoplasy and phenotypic plasticity [17,18]. By using this technique, the illegal trading of protected and endangered wildlife species could also be checked [19,20]. It is also used to identify the species origin in commercially processed food [21]. This is being used as a forensic tool in food safety to check the labeling, substitutions, adulteration, genetically modified organisms, food contamination, and food poisoning [22–31]. It could also be used in identifying seafood that causes health issues in human [32].
India is one of the major exporters of seafood. Any fraudulent substitution in the export is considered as a threat to our economy. There is a possibility of mixing of low priced meat in the place of high priced one. The problem lies in the fact that mislabeled meat is sold at the same price as that of the original meat and the inferior quality meat can cause allergies. In addition to getting substandard foods, consumers are at risk of getting sick as well. The food processing industry must mention the scientific name of the species packed and marketed. In case of any forgery, DNA-based identification always provides a helping hand to identify the exact species. Therefore, in the present study, the frozen shrimp/prawn meat packets marketed by four different leading export companies were chosen to check for any mislabeling or fish fraud. Actually, the aim of the present study was to identify the exact shrimp/prawn species, which have been packed in these company products, by applying the DNA barcoding of mt-COI gene. In fact, this gene was sequenced and compared with sequences available in the NCBI database for similarity check. This method of identification is absolutely trustworthy and thus it is recommended for all food processing and aquaculture industries.
2. MATERIALS AND METHODS
2.1. Sample procurement
The frozen shrimp were obtained from the Nilgiri’s Super Market at R.S. Puram, Coimbatore, India, and a local marketing factory located at Saibaba Colony, Coimbatore, India (Plate 1). The samples belonged to four major food processing brands and exporters in India. They are Sumeru, Innovative Foods Limited, Cochin, India (Sample-A, unlabeled independent species packet), Buffet (Sample-B, unlabeled independent species packet), Cambay Tiger, Mumbai, India (Sample-C, labeled independent species packet), and Britte Seafood, Britto Seafood Exports Pvt. Ltd. (Samples D, E, and F unlabeled mixed-species packet). The sample pockets contained only small-sized prawn meat. The collected samples were stored in a deep freezer. They were subjected to molecular analysis.
2.2. Molecular analysis
2.2.1. DNA isolation
The shrimp meats were subjected to isolation of genomic DNA by adopting the method of Sambrook et al. [33]. Agarose gel electrophoresis (1% AGE) was performed and the genomic DNA was viewed under a gel documentation system (Medicare, India).
2.2.2. DNA amplification
Amplification of DNA of mt-COI gene was done using a Thermo Cycler (Applied Biosystem, USA) with universal primers (LCO1490 & HCO2198) [34]. These primers have worked well with crustacean [35–39]. The amplification process is as given below: total volume, 100 μl (3 μl of DNA template; 1 μl of each primer; 25 μl of 2× PCR Master Mix (MBI Fermentas), and 70 μl of water. The PCR condition was as follows: pre-running (at 95°C for 8 minutes); denaturation (at 95°C for 35 cycles of 30 seconds each); annealing (at 57°C for 30 seconds); extension (at 72°C for 1 minutes); and final extension (at 72°C for 8 minutes). 2% AGE was performed to resolve the amplified sequence of mt-COI gene.
2.2.3. DNA sequencing
Sequencing was done with the following conditions: the reaction mixture (20 μl total volume) containing template DNA (3 μl), primers (3.2 pM/μl of forward (0.50 μl) and reverse (0.50 μl), 5× big dye sequencing buffer (2 μl) and 2.5× ready reaction premix (4 μl, containing Tris-HCL, (pH, 9.0) and MgCI2) and DNase-RNase free water (10 μl). The PCR sequencing condition was as follows: pre-running at 96°C (25 cycles for 1 second each); denaturation at 96°C (25 cycles for 10 seconds each); annealing at 50ºC (25 cycles for 5 seconds each); and elongation at 60ºC (30 cycles of 4 minutes each). After completion of the PCR process, the samples were processed for ethanolic precipitation: the samples were transferred to 96-well microtiter plate, to each well 125 mM EDTA (Ethylene Diamine Tetra-Acetic Acid) (5 μl) and ice cold 100% ethanol (60 μl) stored at −20ºC were added, the plate was sealed and mixed by vortexing for 20–30 seconds and incubated at room temperature for 15 minutes. The plate was spun at 3,000×g at 4ºC for 30 minutes. The supernatant was carefully removed and again spun at 180×g for 1 minute. 60 μl ice cold 70% ethanol (stored at −20ºC) was added to the pellet and centrifuged at 1,650×g at 4ºC for 15 minutes. The plate was inverted again and spun up to 180×g for 1 minute. The samples were re-suspended in Hi-Di formamide (10 μl) and incubated at room temperature for 15 minutes. The re-suspended samples were transferred to the appropriate wells of the sample plate. The samples were denatured with snap chill at 95ºC for 5 minutes and sequenced. The data were analyzed using ABI 3500 XL Genetic Analyzer (Chromous Biotech, Bangalore, India).
2.2.4. Sequence analysis
The sequences were aligned pairwise using CAP3. The similar sequences deposited in the NCBI database were retrieved, internal stop codons were removed through BLAST (Basic Local Alignment Search Tool). The reading frameshift within the sequence was identified and trimmed using ORF finder. The sequences were authenticated with GenBank (L. vannamei, MK792401; Fenneropenaeus merguiensis, MK792399; P. monodon, MK792400; Fenneropenaeus indicus, MK792402; Penaeus semisulcatus, MK792403; and P. monodon, MK792404). The other molecular analysis, such as multiple sequence alignment and highlighting of identical, similar, and variable sites of amino acids was done through the software T-Coffee and multiple align show (MAS), respectively. The phylogenetic information, AT and GC biases, nucleotide divergence (K2P model) [40], and reconstruction of the phylogenetic tree were performed using MEGA (Molecular Evolutionary Genetics Analysis) software (v.6.01).
 | Plate 1: Collected frozen shrimps of various brands. (A) Sumeru, Innovative Foods Limited, Cochin, India (unlabeled independent species packet); (B) Buffet (unlabeled independent species packet); (C) Cambay Tiger, Mumbai, India (labeled independent species packet); (D, E, and F), Britte Seafood, Britto Seafood Exports Pvt. Ltd. (unlabeled mixed-species packet). [Click here to view] |
3. RESULTS AND DISCUSSION
3.1. Genomic DNA and its amplification
The isolated genomic DNA from shrimp samples showed >10 kb nucleotides (Plate 2) and their respective PCR amplified products showed >500 bp length (Fig. 1). Actually, the aligned sequences have 546, 770, 770, 700, 630, and 552 bp for samples A, B, C, D, E, and F, respectively. These sequences showed 99%–100% similarity with the retrieved sequences (Table 1). The sample-A showed 99% similarity with F. merguiensis from Thailand, sample-B showed 100% similarity with P. monodon from Thailand, sample-C showed 98% similarity with L. vannamei from China, sample-D showed 100% similarity with F. indicus from Tanzania, sample-E showed 100% similarity with P. semisulcatus from Sri Lanka, and sample-F showed 100% similarity with P. monodon from India.
3.2. Amino acid residues
These sequences showed 112 identical, 53 similar, and 595 variable amino acids. Therefore, the variable amino acid sites were higher than that of the identical and similar amino acid residues (Table 2 and Fig. 2). This indicated that these shrimp species are genetically discriminated.
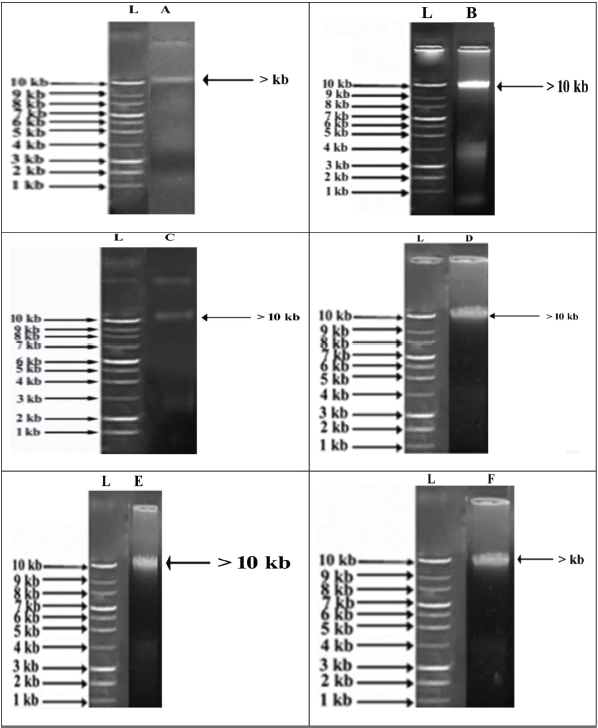 | Plate 2: Genomic DNA of shrimp on AGE (1%). L, 1 kb ladder, shrimp taken from the unlabeled independent packet (A, F. merguiensis; B, P. monodon), labeled independent packet (C, L. vannamei), and unlabeled mixed packet (D, F. indicus; E, P. semisulcatus; F, P. monodon). [Click here to view] |
3.3. Nucleotide composition
The base composition of mt-COI gene sequences varied among these species. Actually, AT bias varied from 61.5% to 67.51% (P. semisulcatus and P. monodon, respectively) and the GC bias varied from 32.9% to 38.6% (P. monodon and P. semisulcatus). In this study, the more AT bias observed suggests that lower abundance of nuclear copies in the mt-DNA of COI gene (NUMTs), known as a pseudogene (Table 3). Similar to the present study, the higher AT bias have also been reported in marine crabs, Portunus sanguinolentus, Charybdis natator, Portunus pelagicus, Portunus trituberculatus, and Travancoriana napaea [41]; freshwater crabs, Spiralothelphusa hydrodroma and Barytelphusa jacquemontii, and freshwater prawns, Macrobrachium rosenbergii, Macrobrachium malcolmsonii, Macrobrachium lamarrei, Macrobrachium lamarrei lamarroids, and Caridina gracilipes [36,37]; freshwater zooplankton, Asplanchna intermedia, Moina micrura, Mesocyclops edax, and Cypris protubera [38,39]. The higher A+T content and lower G+C content have also been reported by Wang et al. [42] in Vitis vinifera.
 | Figure 1: Amplified mt-COI gene product on AGE (2%). Shrimp taken from unlabeled independent packet (A, F. merguiensis; B, P. monodon), labeled independent packet (C, L. vannamei), and unlabeled mixed packet (D, F. indicus; E, P. semisulcatus; F, P. monodon). [Click here to view] |
3.4. Nucleotide divergence
The mean inter species divergence of these frozen shrimp samples were 1.709 with a maximum of 3.401 (between F. merguiensis and P. monodon) and a minimum of 0.434 (between L. vannamei and F. indicus (Table 4). However, the divergent value was >3% in the following two combinations, F. merguiensis (A) versus P. monodon (B) and P. monodon (B) versus L. vannamei (C), and the intraspecies divergence of P. monodon also showed >3% which may occur due to the geographical variation of the species.
3.5. Phylogenetic tree topology
The phylogenetic tree topology of frozen shrimp samples appeared with three clades. At the base, F. indicus formed a single distinct clade and P. semisulcatus also formed a single distinct clade. The rest of the four species, F. merguiensis, L. vannamei, P. monodon (taken from mixed packet), and P. monodon (taken from an independent packet) formed as a cluster in another clade (Fig. 3). Thus, the phylogenetic tree topology indicates clear discrimination between these frozen shrimp samples.
The Telegraph Calcutta reports that more than one-fifth of the seafood sold in markets and restaurants is mislabeled [43]. Mixing of fish species can lead to that with contaminants, toxins, and allergens to be consumed making the consumer sick [44]. The study stresses the importance of labeling marketed seafood, be it in whole meat form or deveined, de-shelled frozen form. The novelty of the work lies in the fact that DNA bar-coding is a suitable tool to exactly pinpoint the species that are marketed in case the manufacturers haven’t labeled the package or in times where we suspect fraudulent substitutions. The samples used in this study consisted of four major food processing brands, out of which only one brand had specified the shrimp that had been packaged (Sample C—L. vannamei). Samples A and B were found to be F. merguiensis and P. monodon, respectively, whereas the packaged frozen meat from Britte Seafood, Britto Seafood Exports Pvt. Ltd. consisted of mixed shrimps D—F. indicus, E—P. semisulcatus, F—P. monodon which was identified using DNA barcoding. These unlabeled packages that are sold in supermarkets with a high market value are sometimes of a fraudulent nature as shrimp meats are mixed. This mixing of species is often due to fraud or even human error. This has an impact on the international markets with meats of high value being substituted with the meat of low economic value [45]. DNA barcoding can be a suitable solution as the species used can be easily and accurately identified and the fraud averted. This study is novel as it combines a good and reliable technology and a common problem in the seafood industry and brings about an effective solution that can be employed. The food processing industries must compulsorily label their products in case if distinguishable parts have been removed and even in packages that are imported or sold in the local markets so that consumers are aware of what they eat.
 | Table 1: BLAST identification of COI gene sequences generated for frozen shrimp. [Click here to view] |
 | Table 2: Number of identical, similar, and variable sites of amino acid residues in the COI gene sequences generated for different subjected frozen shrimp species. [Click here to view] |
 | Figure 2: Multiple sequence alignment of COI gene sequences generated for subjected frozen shrimp species (A, F. merguiensis; B, P. monodon; C, L. vannamei D, F. indicus; E, P. semisulcatus; F, P. monodon). An alignment is formatted by using multiple align show (MAS) with colored background and a consensus setting of 100%. Identical residues are indicated by amino acid color and similar residues are black in color. Gaps and other residues are given in white background. [Click here to view] |
 | Table 3: Nucleotide composition percentage of independent subjected frozen shrimp. [Click here to view] |
 | Table 4: Nucleotide divergence of frozen shrimp. [Click here to view] |
 | Figure 3: Phylogenetic tree topology of subjected shrimp species. [Click here to view] |
4. CONCLUSION
This study concluded that the unknown shrimp species in frozen meat samples were clearly identified and discriminated through DNA barcoding of mt-COI gene, a tool for identifying the unknown seafood.
REFERENCES
1. Nagalakshmi K, Annam P, Venkateshwarlu G, Pathakota GB, Lakra WS. Mislabeling in Indian seafood: An investigation using DNA barcoding. Food Control 2015;59:196–200.
2. Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. DNA barcoding Australia’s fish species. Philos Trans R Soc Lond B 2005;360:1847–57.
3. Van Leeuwen SP, Van Velzen MJ, Swart CP, van der Veen I, Traag WA, de Boer J. Halogenated contaminants in farmed salmon, trout, tilapia, pangasius and shrimp. Environ Sci Technol 2009;43:4009–15.
4. Marko PB, Lee SC, Rice AM, Gramling JM, Fitzhenry TM, McAlister JS. Fisheries: mislabeling of a depleted reef fish. Nature 2004;430(6997):309–10.
5. Ward RD. Genetics in fisheries management. Hydrobiologia 2000;420:191–201.
6. Warner K, Timme W, Lowell B, Hirschfield M. Oceana study reveals seafood fraud nationwide, 2013. Available via http://oceana.org/reports/oceana-study-reveals-seafood-fraud-nationwide.
7. Smith PJ, McVeagh SM, Steinke D. DNA barcoding for the identification of smoked fish products. J Fish Biol 2008;72:464–71.
8. Wong EHK, Hanner RH. DNA barcoding detects market substitution in North American seafood. Food Res Inter 2008;4:828–37.
9. Holmes BH, Steinke D, Ward RW. Identification of shark and ray fins using DNA barcoding. Fish Res 2009;95:280–8.
10. Ward RD, Holmes BH, White WT, Last PR. DNA barcoding Australasian Chondrichthyans: results and potential uses in conservation. Mar Fresh Res 2008;59:57–71.
11. USDHHS. The fish list” FDA guide to acceptable market names for food fish sold in interstate commerce. Department of Health and Human Service, FDA Center for Food Safety and Applied Nutrition, available from Sup. Documents, US Govt. Printing Office, Washington, DC, 1988.
12. Jacquet JL, Pauly D. Trade secrets: renaming and mislabeling of seafood. Mar Policy 2008;32:309–18.
13. Ogden R. Fisheries forensics: the use of DNA tools for improving compliance, traceability and enforcement in the fishing industry. Fish Fisheries, 2008;9:462–72.
14. Rubinoff D. Utility of mitochondrial DNA Barcodes in species conservation. Conserv Biol 2006;20:1026–33.
15. Filonzi L, Chiesa S, Vaghi M, Marzano FN. Molecular barcoding reveals mislabeling of commercial fish products in Italy. Food Res Int 2010;43:1383–8.
16. Wong LL, Peatman E, Lu J, Kucuktas H, He S, Zhou C. DNA barcoding of catfish: Species authentication and phylogenetic assessment. PLoS One 2011;6(3):e17812.
17. Pegg GG, Sinclair B, Briskey L, Aspden WJ. MtDNA barcode identification of fish larvae in the southern Great Barrier Reef, Australia. Sci Mar 2006;70:7–12.
18. Vences M, Thoma M, Bonett RM, Vieites DR. Deciphering amphibian diversity through DNA barcoding: chances and challenges. Philos Trans R Soc B 2005;360:1859–68.
19. Baker CS, Lento GM, Cipriano F, Palumbi SR. Predicted decline of protected whales based on molecular genetics monitoring of Japanese and Korean markets. Proc R Soc B 2000;267:1191–9.
20. Shivji M, Clarke S, Pank M, Natanson L, Kohler N, Stanhope M. Genetic identification of pelagic shark body parts for conservation and trade monitoring. Conserv Biol 2002;16:1036–47.
21. Dawnay N, Ogden R, McEwing R, Carvalho GR, Thorpe RS. Validation of the barcoding gene COI for use in forensic genetic species identification. Forensic Sci Int 2007;173:1–6.
22. Tanabe S, Miyauchi E, Muneshige A, Mio K, Sato C, Sato M. PCR method of detecting pork in foods for verifying allergen labelling and for identifying hidden pork ingredients in processed foods. Biosci Biotechnol Biochem 2007;71:1663–7.
23. Hsieh YHP. Species substitution of restaurant fish entrees. J Food Qual 1998;21:1–11.
24. Teletchea F, Maudet C, Hanni C. Food and forensic molecular identification: update and challenges. Trends Biotech 2005;23:359–66.
25. Ronning SB, Rudi K, Berdal KG, Holst-Jensen A. Differentiation of important and closely related cereal plant species (Poaceae) in food by hybridization to an oligonucleotide array. J Agric Food Chem 2005;53:8874–80.
26. Hsieh YW, Shiu YC, Cheng CA, Chen SK, Hwang DF. Identification of toxin and fish species in cooked fish liver implicated in food poisoning. J Food Sci 2002;67:948–52.
27. Ardura A, Linde AR, Moreira JC, Garcia-Vazquez E. DNA barcoding for conservation and management of Amazonian commercial fish. Biol Conserv 2010;143:1438–43.
28. Hanner R, Becker S, Ivanova NV, Steinke D. FISH-BOL and seafood identification: geographically dispersed case studies reveal systemic market substitution across Canada. Mitochon DNA 2011;22:106–22.
29. Lowenstein JH, Amato G, Kolokotronis SO. The real maccoyii: identifying tuna sushi with DNA barcodes—contrasting characteristic attributes and genetic distances. PLoS One 2009;4:e7866.
30. Torres RA, Feitosa RB, Carvalho DC, Freitas MO, Hostim-Silva M, Ferreira BP. DNA barcoding approaches for fishing authentication of exploited grouper species including the endangered and legally protected goliath grouper Epinephelus itajara. Sci Mar 2013;77:409–18.
31. Vandamme SG, Griffiths AM, Taylor SA, Di Muri C, Hankard EA, Towne JA, Watson M, Mariani S. Sushi barcoding in the UK: another kettle of fish. Peer J 2016;4:e1891.
32. Lowenstein JH, Burger J, Jeitner CW, Amato G, Kolokotronis SO, Gochfeld M. DNA barcodes reveal species-specific mercury levels in tuna sushi that pose a health risk to consumers. Biol Lett 2010;6:692–5.
33. Sambrook J, Fritschi EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY, 1989.
34. Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome oxidase subunit I from diverse metazoan invertebrates. Mol Mar Bio Biotechnol 1994;3:294–9.
35. Rajkumar G, Bhavan PS, Udayasuriyan R and Vadivalagan C. Molecular identification of shrimp species, Penaeus semisulcatus, Metapenaeus dobsoni, Metapenaeus brevicornis, Fenneropenaeus indicus, Parapenaeopsis stylifera and Solenocera crassicornis inhabiting in the coromandel coast (Tamil Nadu, India) using MT-COI gene. Int J Fish Aqua Stud 2015;2(4):96–106.
36. Udayasuriyan R, Bhavan PS, Vadivalagan C, Rajkumar G. Efficiency of different COI markers in DNA barcoding of freshwater prawn species. J Entomol Zool Stud 2015;3(3):98–110.
37. Bhavan PS, Umamaheswari S, Udayasuriyan R, Rajkumar G, Amritha H, Saranya K. Discrimination of two freshwater crabs Spiralothelphusa hydrodroma and Barytelphusa jacquemontii and one mangrove crab Neosarmatium asiaticum by DNA barcoding of MT-COI gene. J Chem Biol Physical Sci 2015;5:1426–40.
38. Bhavan PS, Udayasuriyan R, Vadivalagan C, Kalpana R, Umamaheswari S. Diversity of zooplankton in four perennial lakes of Coimbatore (India) and molecular characterization of Asplanchna intermedia, Moina micrura, Mesocyclops edax and Cypris protubera through mt-COI gene. J Entomol Zool Stud 2016;4(2):183–97.
39. Bhavan PS, Udayasuriyan R, Kalpana R, Gayathri M. Molecular identification and characterization of few crustacean zooplankton species by mt-COI gene. J Biol Nat 2017;7(1):1–23.
40. Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16:111–20.
41. Umamaheswari S, Bhavan PS, Udayasuriyan R, Vadivalagan C, Kalpana R. Discrimination of four marine crabs and one freshwater crab through mt-COI gene. J Entomol Zool Stud 2016;4(5):766–82.
42. Wang Y, Liu X, Ren C, Zhong G, Yang L, Li S. Identification of genomic sites for CRISPR/Cas9-based genome editing in the Vitis vinifera genome. BMC Plant Biol 2016;21:16–96.
43. Something fishy! One-fifth of the seafood sold in markets is mislabeled. Available via https://www.firstpost.com/india/something-fishy-one-fifth-seafood-sold-markets-mislabelled-2376576.html (Accessed 3 August 2015).
44. Health Risks of Seafood Fraud. Available via https://usa.oceana.org/health-risks-seafood-fraud
45. Stawitz CC, Siple MC, Munsch SH, Lee Q, Derby SR. Financial and ecological implications of global seafood mislabeling. Conserv Lett 2017;10(6):681–9.