1. INTRODUCTION
Acid phosphatases (orthophosphoric-monoester phosphohydrolase, EC 3.1.3.2) are well known for hydrolysis of phosphomonoesters [1]. These enzymes exhibited many biological roles as metabolism regulation, cellular signal transduction pathways [2], and defence against herbivorous insects [3]. The action of acid phosphatases on phosphorylated compounds transforms organic phosphate into a soluble inorganic form, available for many cellular functions in living organisms [4,5]. Knowing the importance of inorganic phosphate in plants, animals, and microorganisms growth, we would understand why phosphatases are widespread in nature [6]. Indeed, phosphorus is an essential element in all metabolic processes and in synthesis of nucleic acids and membranes [7].
Like most biocatalysts, acid phosphatases are applied as valuable tools for various biotechnological and bioindustrial purposes [8]. Their uses in environmental bioremediation of polluted soils (e.g., removal of heavy metals) has been reported by several studies [9–11]. In the context of sustainable agriculture, phosphatases could increase soils fertility by phosphate solubilization, and thus, minimize the chemical fertilizer application [12]. Moreover, phytate-specific phosphatases called phytases were added to the diet of monogastric animals to improve phosphate absorption from phytate (myo-inositol hexakisphosphate) which is the major storage form of phosphate in plant seeds [13]. Another key area of acid phosphatases interest in biotechnology is their exploitation to produce phosphoconjugates as nucleotides. These nucleotides are highly sought after in the food and pharmaceutical industries for their taste and flavor potentialities [14–16]. However, temperature is a key variable in bioprocesses using enzymes [17]. Due to the fact that many industrial processes require stable and sturdy biocatalysts, it is essential to know their thermal stability and thermodynamic properties. According to Ward and Moo [18], temperature is the best optimized physical variable in chemical reactions. Generally, thermostable enzymes have several advantages in terms of strength, reactivity, and process yield. Hence, this could translate the ability of enzyme to maintain maximum activity over time, since enzyme stability represents a critical point in most bioprocesses [19,20].
To our knowledge, despite the various applications mentioned above, very few reports detailed thermodynamic characterization and thermal stability of acid phosphatases. In this paper, we report on the thermostability parameters of non-specific acid phosphatases BLsAP1, BLsAP2, RLsAP1, and RLsAP2 [21,22], isolated from edible seeds of Lagenaria siceraria (Molina) Standl., an oleaginous cucurbit largely consumed in Sub-Saharan Africa [23].
2. MATERIALS AND METHODS
2.1. Enzymes
The phosphatases BLsAP1, BLsAP2, RLsAP1, and RLsAP2 (EC 3.1.3.2) originated from the purified enzyme collection of “Laboratoire de Biotechnologies, UFR Biosciences, Université Felix Houphouët-Boigny (Abidjan, Côte d’Ivoire).” BLsAP1 and BLsAP2 were isolated from seeds of L. siceraria blocky-fruited cultivar, while RLsAP1 and RLsAP2 were isolated from seeds of the round-fruited cultivar as described previously by Koffi et al. [21,22].
2.2. Enzyme assay
The acid phosphatase activity was carried out in a total volume of 250 μl, containing 100 mM sodium acetate buffer (pH 5.6), substrate (pNPP, 5 mM), and the enzyme solution (25 μl). The reaction mixture was incubated at 37°C for 10 minutes. Then, 2 ml of Na2CO3 2% (w/v) were added to stop the reaction and absorbances were measured at 410 nm by using a spectrophotometer GENESIS 5. para-Nitrophenol (pNP) was used as standard.
One unit of activity was defined as the amount of enzyme that hydrolyzes 1 μmol of substrate per min under the enzyme assay conditions. The specific activity was expressed as a unit of activity per mg of protein.
2.3. Protein estimation
Protein concentrations and elution profiles from chromatographic columns were determined by the Folin method [24]. Bovine serum albumin (BSA) was used as the standard protein.
2.4. Thermal Inactivation
The thermal inactivation of each purified acid phosphatase was determined at various constant temperatures from 55°C for RLsAP2 (its optimum temperature) and 60°C for BLsAP1, BLsAP2, and RLsAP1 (their optimum temperature) to 80°C. Enzymes in sodium acetate buffer (pH 5.6) were exposed to each temperature for a period of 5 to 60 minutes. Then, aliquots were withdrawn at intervals (5 minutes) and immediately cooled at 4ºC for 20 minutes. Residual activities, determined at 37°C under the standard enzyme assay conditions, were expressed as percentage of activity of zero-time control of untreated enzymes.
2.5. Residual activity
where A and A0 are the activities of the treated and untreated enzyme solution, respectively.
2.6. Kinetic data analysis
2.6.1. Denaturation constants of the enzyme fraction k
Thermal inactivation of each phosphatase can be described by a first-order kinetic model [25]. The integral effect of an inactivation process at constant temperature, where the inactivation rate constant is independent of time, is given in Equation /(1) as follows:
where At is the residual enzyme activity at time t (minutes), A0 is the initial enzyme activity, k (minute−1) is the inactivation rate constant at a given condition. k values were obtained from the regression line of ln (At /Ao) versus time as slope.
2.6.2. Determination of half-life of the enzyme solution t1/2
The t1/2 was obtained from the following equation:
2.6.3. Calculation of activation Energy Ea
The Ea was obtained from the Arrhenius equation:
where k is the first-order rate constant of thermal inactivation of the enzyme activity, obtained through the slopes of the plots described above.
2.6.4. Calculation of D values of the enzyme fractions
D-value is defined as the time needed, at a constant temperature, to reduce the initial enzyme activity (A0) by 90 % (or decimal reduction time or time required to preincubate the enzyme at a given temperature to maintain 10% residual activity). For first-order reactions, the D-value is directly related to the rate constant k (Eq. 2) [26,27]:
2.6.5. Calculation of z values of the enzyme fractions
The z value (°C) is the temperature increase needed to induce a 10-fold reduction in D value [26]. This z value follows the Eq. (6):
where T1 and T2 are the lower and higher temperatures in °C or K. Then, D1 and D2 are D values at the lower and higher temperatures in minute, respectively.
The z values were determined from the linear regression of log (D) and temperature (T).
2.6.6. Thermodynamic parameters
The enthalpy of reaction (ΔH#), which is the amount of energy required to bring the enzyme to the activated state for the subsequent denaturation at a given temperature, Gibbs free energy (ΔG#) and the entropy (ΔS#) were calculated through the following equations [28,29].
where ΔH# = Change in enthalpy; Ea = activation energy of inactivation; R = general gas constant; T = temperature.
where ΔG# = free energy change of inactivation; h = Plank’s constant; Kb = Bolztman constant.
3. RESULTS AND DISCUSSION
As reported by Koffi et al. [21,22], acid phosphatases purified from L. siceraria seeds were mesophilic enzymes. Thus, BLsAP1, BLsAP2, and RLsAP1 showed maximal hydrolytic activities at 60°C, while RLsAP2 has an optimum activity at 55°C. The present study shows that when submitted to heat treatment, these enzymes are completely inactivated between 75°C and 80°C after 30 minutes (data not shown). It is well known that enzymes are proteins with three-dimensional structure which are essential for their activities [30]. It seems that after a long-term exposure to heat, this enzyme three-dimensional structure was destabilized and then, inactivated. So, a biocatalyst capable to resist against thermal unfolding in absence of substrate is categorized as thermostable, while an enzyme molecule able to catalyze reactions (that means substrate presence) at high temperatures (beyond 70°C) is qualified of thermophilic [31].
The thermal inactivation of the four studied acid phosphatases represented in Figure 1 shows that these enzymes followed a first-order kinetic. This is explained by the logarithmic linear relationship between the phosphatase activities and time of treatment for the temperatures ranged from 55°C to 80°C (Fig. 1). Similar results have been reported for α-amylases from sprouted pearl millet [32]. Moreover, this linear appearance for the graph ln (At /A0) suggests that each reaction mixture contains only one enzyme that catalyzes the substrate hydrolysis. Indeed, the combined activities of several phosphatases (in the same reaction mixture) would have resulted in a curve with many phases instead of a straight line.
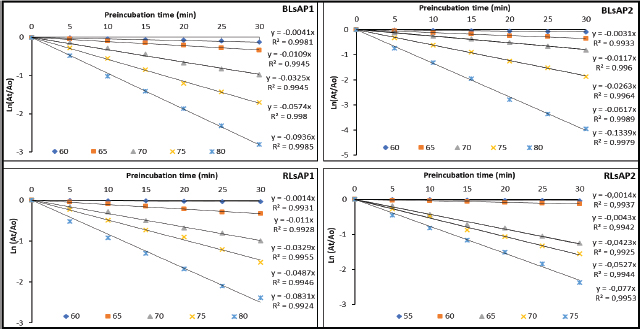 | Figure 1: First order thermal inactivation of acid phosphatases BLsAP1, BLsAP2, RLsAP1, and RLsAP2 isolated from L. siceraria seeds at different temperatures. A0 is the initial enzymatic activity and At the activity at each holding time. [Click here to view] |
From the slope of the graphs in Figure 1, the inactivation rate constants (k) were estimated. Then, k values and half-life (t1/2) of studied acid phosphatases are presented in Table 1. These results show that k values increase with increasing pre-incubation temperature for the four enzymes. This confirms the instability of these enzymatic proteins at high temperatures as reported by Marangoni [33]. This author argues that higher value of k constant means that the enzyme was less thermostable. As concerned the half-life of acid phosphatases from L. siceraria seeds, they appear to be too longer at optimum temperatures, then they decrease beyond. At their optimum temperatures, acid phosphatases BLsAP1, BLsAP2, RLsAP1, and RLsAP2 showed half-lives of 169.06, 223.60, 495.11, and 495.10 minutes, respectively. This result suggests that acid phosphatases RLsAP1 and RLsAP2 isolated from the round-fruited cultivar of L. siceraria were more thermally stable than those from the blocky-fruited cultivar seeds. However, the half-lives found in this study were higher compared to half-life obtained for beta-glucosidase from land crab (Cardisoma armatum) digestive juice which was of 169.02 minutes at 60°C [34].
Figure 2 depicts the effect of temperature on D values for inactivation of acid phosphatases isolated from L. siceraria edible seeds. The D values, which are the time required to reduce the enzyme activities by 90%, corroborate results obtained for half-life. Indeed, for the four studied acid phosphatases, D values decreased very sharply with increasing temperature. This is the proof that the inactivation time of the studied enzymatic molecules is shortened rapidly at high temperatures. As shown in Table 2, at their optimum temperature, D values obtained were of 561.71, 742.90, 1645.00, and 1645.03 minutes for acid phosphatases BLsAP1, BLsAP2, RLsAP1, and RLsAP2, respectively. It is noteworthy that at 60°C, the D value for RLsAP1 inactivation was, respectively, 2.93 and 2.21 times higher than the corresponding value for inactivation of BLsAP1 and BLsAP2. This indicates probably the higher thermal stability of RLsAP1. Otherwise, for this acid phosphatase, the decimal time is reduced about 8-fold when the optimal temperature increases by 5°C (from 1,645.00 to 209.36 minutes). D values (ranged from 258.7 ± 11.4 minutes to 0.41 ± 0.05 minutes in buffer solution) reported for alkaline phosphatase (EC 3.1.3.1) from calf intestine were lower than those obtained in this study [35].
 | Table 1: k values and half-lives of acid phosphatases BLsAP1, BLsAP2, RLsAP1, and RLsAP2 isolated from L. siceraria seeds. [Click here to view] |
.png) | Figure 2: Effect of temperature on D values for inactivation of acid phosphatases BLsAP1, BLsAP2, RLsAP1, and RLsAP2 isolated from L. siceraria seeds. [Click here to view] |
The temperature resistance values (temperature increase needed for a 10-fold decrease of D) were calculated and results are shown in Table 2. The corresponding z values were 14.53°C, 12.52°C, 11.92°C, and 10.94°C for the inactivation of acid phosphatases BLsAP1, BLsAP2, RLsAP1, and RLsAP2, respectively. These results are consistent with values obtained for alkaline phosphatases in raw milk (11.8°C) by Fadilo?lu et al. [36] and in whole milk (12.27°C) by St?nciuc et al. [35]. Previously, Blel et al. [37], Claeys [38], Levieux et al. [39], and Marchand et al. [40] reported z values ranging from 5°C to 8°C (for thermal inactivation of alkaline phosphatases) lower than z values obtained in this study.
The slopes of Arrhenius plots in Figure 3 allow us to calculate the activation energy (Ea) of each enzyme. Activation energy is a minimum amount of energy required for the native enzyme in order to initiate the inactivation or denaturation process. This activation energy can be seen as the energy barrier that molecules need to cross to be able to react [41]. Beyond the activation energy, the enzyme is denatured and cannot refold to the native form as reported by Siddiqui et al. [28]. The Ea values of acid phosphatases from L. siceraria seeds were ranged from 155.08 (for BLsAP1) to 200.55 kJ/mol (for RLsAP2) (Table 2). For example, it means that for acid phosphatase BLsAP1, it was necessary to absorb 155.08 kJ from the external medium to start its inactivation at temperatures between 60°C and 80°C. These values were higher than those found (92.4 and 81.243 kJ/mol) for α-amylases from sprouted pearl millet [32]. The relative high values obtained for activation energies suggest that important energy amounts must be supplied to the studied enzyme in order to initiate the denaturation process. It means that these acid phosphatases are relatively thermostable.
The activation energy value enabled the determination of enthalpy, entropy, and free energy of activation of the four enzymes as shown in Table 3. Enthalpy (ΔH#) is the total amount of energy required to bring the enzyme from the ground state to the activated intermediate. In this study, enthalpy values (ΔH#) decreased as the temperature increased (Table 3). The obtained ΔH# average values were 152.23, 177.04, 186.82, and 197.74 kJ.mol-1 for BLsAP1, BLsAP2, RLsAP1, and RLsAP2, respectively. According to Lai and Tabatabai [42], a greater amount of stretching, squeezing, or breaking of chemical bonds required to reach the transition state corresponds to a larger value of ΔH#. Among the four studied acid phosphatases, it appears that RLsAP2 (which shows the much higher ΔH#) would undergo the most conformational change during thermal inactivation as reported by Marin et al. [43]. Being well known that ΔH# and Ea are directly related, the high enthalpy (ΔH#) change for RLsAP1 and RLsAP2 indicates that these enzymes need greater energy for their thermal denaturation [44]. So, the positive value of the enthalpy change (ΔH#) would suggest the endothermic nature of these enzymatic reactions.
 | Table 2: D, Z and Ea values for thermal inactivation of phosphatases BLsAP1, BLsAP2, RLsAP1, and RLsAP2 from L. siceraria seeds. [Click here to view] |
The positive values of the obtained entropy (ΔS#) and free energy (ΔG#) corroborate the fact that the studied acid phosphatases underwent a large change in conformation [35]. Activation entropy (ΔS#) is defined as the degree of disorder in the transition complex, while Gibbs activation free energy (ΔG#) represents the spontaneity of a reaction and the energy required for the enzyme-substrate complex to breakdown into a catalytic reaction product [29]. the obtained ΔS# values of acid phosphatases from L. siceraria seeds were ranged from 175.34 (for BLsAP1) to 313.35 J.mol-1.K-1 (for RLsAP2) (Table 3). The positive values obtained for entropy changes (ΔS#) would translate a high disorder at the transition state, which is not the case in the native state the native state. Also, it would indicate that aggregation processes are not significant.
The Gibbs free energy of activation (ΔG#) represents a good index to measure stability of biocatalyst [45]. The reaction is said spontaneous when the Gibbs free energy is low or negative. In this study, positive ΔG# values obtained imply that thermal inactivation reactions of acid phosphatases from L. siceraria were not spontaneous. So, the decrease of ΔG# for all studied acid phosphatases with increasing temperature suggests lower stability at high temperature [46,47].
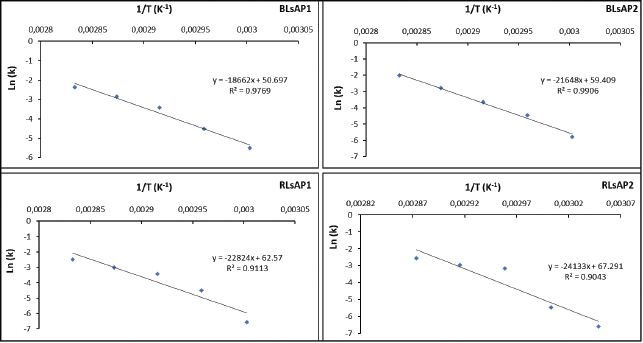 | Figure 3: Arrhenius plot for thermal inactivation of acid phosphatases BLsAP1, BLsAP2, RLsAP1, and RLsAP2 isolated from L. siceraria seeds. [Click here to view] |
 | Table 3: Thermodynamic parameters of acid phosphatases BLsAP1, BLsAP2, RLsAP1, and RLsAP2 from L. siceraria seeds. [Click here to view] |
4. CONCLUSION
Investigation on thermal inactivation of acid phosphatases from L. siceraria seeds indicated a first-order kinetics for the four enzymes. They showed high activation energies (Ea) and change in enthalpy (ΔH#) suggesting that these acid phosphatases are relatively thermostable since they need important energy amounts before triggering the inactivation process. The comparison of the rate constants, half-lives, and D values revealed a better thermostability of the two acid phosphatases (RLsAP1 and RLsAP2) isolated from the round-fruited cultivar of L. siceraria.
REFERENCES
1. Vincent JB, Crowder MW, Averill BA. Hydrolysis of phosphate monoester: a biological problem with multiple chemical solutions. Trends Biochem Sci 1992;17:105–10. CrossRef
2. Thaller MC, Berlutti F, Schippa S, Iori P, Passariello C, Rossolini GM. Heterogenous patterns of acid phosphatase containing low-molecular-mass polypeptides in members of the family Enterobacteriaceae. Int J System Bacteriol 1995;45:255. CrossRef
3. Liu Y, Ahn JE, Datta S, Salzman RA, Moon J, Huyghues-Despointes B, et al. Arabidopsis vegetative storage protein is an anti-insect acid phosphatase. Plant Physiol 2005;139:1545–56. CrossRef
4. Yadav J, Verma JP, Tiwari KN. Plant growth promoting activities of fungi and their effect on Chickpea plant growth. Asian J Biol Sci 2011;4(3):291–9. CrossRef
5. Anand A, Srivastava PK. A Molecular description of acid phosphatase. Appl Biochem Biotechnol 2012;167(8):2174–97. CrossRef
6. Zhang GQ, Chen QJ, Sun J, Wang HX, Han CH. Purification and characterization of a novel acid phosphatase from the split gill mushroom Schizophyllum commune. J Basic Microbiol 2013;53:1–8. CrossRef
7. Ehsanpour AA, Amini F. Effect of salt and drought stress on acid phosphatase activities in alfalfa (Medicago sativa L.) explants under in vitro culture. Afr J Biotechnol 2003;2:133–5. CrossRef
8. Mehra P, Pandey B K, Giri J. Improvement in phosphate acquisition and utilization by a secretory purple acid phosphatase (OsPAP21b) in rice. Plant Biotechnol J 2017;15:1054–67. CrossRef
9. Basnakova G, Stephens ER, Thaller MC, Rossolini GM, Macaskie LE. The use of Escherichia coli bearing a pho N gene for the removal of uranium and nickel from aqueous flows. Appl Microbiol Biotechnol 1998;50:266–72. CrossRef
10. Macaskie LE, Bonthrone KM, Yong P, Goddard DT. Enzymically mediated bioprecipitation of uranium by a Citrobacter sp.: a concerted role for exocellular lipopolysaccharide and associated phosphatase in biomineral formation. Microbiology 2000;146:1855–67. CrossRef
11. Misra CS, Appukuttan D, Kantamreddi VS, Rao AS, Apte SK. Recombinant D. radiodurans cells for bioremediation of heavy metals from acidic/neutral aqueous wastes. Bioengineered Bugs 2012;3:44–8. CrossRef
12. Behera BC, Yadav H, Singh SK, Mishra RR, Sethi BK, Dutta SK, et al. Phosphate solubilization and acid phosphatase activity of Serratia sp. isolated from mangrove soil of Mahanadi river delta, Odisha, India. J Genet Eng Biotechnol 2017;15:169–78. CrossRef
13. Greiner R, Konietzny U. Phytase for food application. Food Technol Biotechnol 2006;44:125–40.
14. Mihara Y, Utagawa T, Yamada H, Asano Y. Phosphorylation of nucleosides by the mutated acid phosphatase from Morganella morganii. Appl Environ Microbiol 2000;66:2811–6. CrossRef
15. Ishikawa K, Mihara Y, Shimba N, Ohtsu N, Kwasaki H, Suzuki E-I, et al. Enhancement of nucleoside phosphorylation activity in an acid phosphatase. Protein Eng 2002;15(7):539–43. CrossRef
16. Koffi DM, Faulet BM, Gonnety JT, Bédikou ME, Kouamé LP, Zoro Bi IA, et al. Novel plant acid phosphatases from the Orphan Crop Lagenaria siceraria (Molina) Standl. for phosphate ester synthesis. Philipp Agric Sci 2012;95(1):14–21.
17. Wasserman B. Thermostable enzyme production. Food Technol 1984;38:78–88.
18. Ward OP, Moo-Young M. Thermostable enzymes. Biotechnol Adv 1988;6(1):39–69. CrossRef
19. Mozhaev VV. Mechanism-based strategies for protein thermostabilization. Trends Biotechnol 1993;11:88–95. CrossRef
20. Illanes A. Stability of biocatalysts. Electron J of Biocatal 1999;2(1): 1–2. CrossRef
21. Koffi DM, Gonnety JT, Faulet BM, Bédikou ME, Kouamé LP, Zoro Bi IA, et al. Biochemical characterization of two non-specific acid phosphatases from Cucurbitaceae (Lagenaria siceraria) edible seeds exhibiting phytasic activity. J Anim Plant Sci 2010;7(3):860–75. CrossRef
22. Koffi DM, Faulet BM, Gonnety JT, Bédikou ME, Kouamé LP, Zoro Bi IA, et al. Biochemical characterization of two acid phosphatases purified from edible seeds of the neglected crop Lagenaria siceraria (Molina) Standl. blocky-fruited cultivar. Philipp Agric Sci 2010;93(3):269–80.
23. Zoro Bi IA, Kouamé KK, Djè Y, Malice M, Baudoin JP: Indigenous cucurbits of Côte d'Ivoire: a review of their genetic resources. Sci Nat 2006;3:1–9.
24. Lowry OH, Rosebrough NJ, Farra L, Randall RJ. Protein measurement with Folin phenol reagent. J Biol Chem 1951;193:265–75.
25. Guiavarc'h YP, Deli V, Van Loey AM, Hendrickx ME. Development of an enzymic time temperature integrator for sterilization processes based on Bacillus licheniformis α-amylase at reduced water content. J Food Sci 2002;67(1):285–91. CrossRef
26. Stumbo CR. Thermobacteriology in food processing. 2nd edition, Academic Press, New York, NY, p 336, 1973.
27. Espachs-Barroso A, Van Loey A, Hendrickx M, Martín-Belloso O. Inactivation of plant pectin methylesterase by thermal or high intensity pulsed electric field treatments. Innov Food Sci Emerg Technol 2006;7(1):40–8. CrossRef
28. Siddiqui KS, Azhar MJ, Rashid MH, Rajoka MI. Stability and identification of active-site residues of carboxymethylcellulases from Aspergillus niger and Cellulomonas biazotea. Folia Microbiol 1997;42(4):312–8. CrossRef
29. Riaz MR, Perveen MR, Javed H, Nadeem, MH Rashi. Kinetics and thermodynamic properties of noval glucoamylase from Humicola sp. Enzyme Microb Technol 2007;41:558–64. CrossRef
30. Petsko GA, Ringe D. Protein structure and function. New Science Press in Association with Sinauer Associates and Blackwell Science, London, UK, p 45, 2004.
31. Georis J, De Lemos Esteves F, Lamotte-Brasseur J, Bougnet V, Devreese B, Giannotta F, et al. An additional aromatic interaction improves the thermostability and thermophilicity of a mesophilic family 11 xylanase: structural basis and molecular study. Protein Sci 2000;9(3):466–75. CrossRef
32. Agbo KU, Okwuenu PC, Ezugwu AL, Eze Soo, Chilaka FC. Thermostability and thermodynamic characterization of sprouted pearl millet α-amylases for its biotechnological applications. Bangladesh J Sci Indus Res 2017;52(3):159–66. CrossRef
33. Marangoni AG. Characterization of enzyme stability. In: Enzyme kinetics: a modern approach, John Wiley & Sons, Inc., Hoboken, NJ, 2003.
34. Ya KC, Konan KH, Gnangui SN, Kouamé LP. Study of thermal stability of beta-glucosidase from the land crab digestive juice (cardisoma armatum): kinetic and thermodynamic analysis. Int J Dev Res 2014;4(9):1836–40.
35. St?nciuc N, Ardelean A, Diaconu V, Râpeanu G, Stanciu S, Nicolau A. Kinetic and thermodynamic parameters of alkaline phosphatase and glutamyl transferase inactivation in bovine milk. Dairy Sci Technol 2011;91(6):701–17. CrossRef
36. Fadilo?lu S, Erkmen O, ?ekero?lu GS. Thermal inactivation kinetics of alkaline phosphatase in buffer and milk. J Food Process Preserv 2006;30:258–68. CrossRef
37. Blel M, Guingamp MF, Gaillard JL, Humbert G. Studies on the thermal sensitivity of γ-glutamyl transpeptidase measured with a modified test procedure and compared with that of alkaline phosphatase and lactoperoxidase in milk. Lait 2002;82:555–66. CrossRef
38. Claeys WL, Ludikhuyze L, Van Loey A, Hendrickx ME. Inactivation kinetics of alkaline phosphatase and lactoperoxidase, and denaturation kinetics of β-lactoglobulin in raw milk under isothermal and dynamic temperature conditions. J Dairy Res 2001;68:95–107. CrossRef
39. Levieux D, Geneix N, Levieux A. Inactivation-denaturation kinetics of bovine milk alkaline phosphatase during mild heating as determined by using a monoclonal antibody-based immunoassay. J Dairy Res 2007;74:296–301. CrossRef
40. Marchand S, Merchiers M, Messens W, Coudijzer K, De Block J. Thermal inactivation kinetics of alkaline phosphatase in equine milk. Int Dairy J 2009;19:763–7. CrossRef
41. Yan J, Pan G, Ding C, Quan G. Kinetic and thermodynamic parameters of β-glucosidase immobilized on various colloidal particles from a paddy soil. Colloids Surf B: Biointerfaces 2010;79:298–303. CrossRef
42. Lai CM, Tabatabai MA. Kinetics parameters of immobilized urease. Soil Biol Biochem 1992;24:225–8. CrossRef
43. Marín E, Sánchez L, Pérez MD, Puyol P, Calvo M. Effect of heat treatment on bovine lactoperoxidase activity in skim milk: kinetic and thermodynamic analysis. J Food Sci 2003;68(1):89–93. CrossRef
44. Bhatti HN, Zia A, Nawaz R, Sheikh MA, Rashid MH, Khalid AM. Effect of copper ions on thermal stability of glucoamylase from Fusarium sp. Int J Agric Biol 2005;7:585–7.
45. Marangoni AG. Enzyme kinetics: a modern approach, John Wiley and Sons, New Jersey, pp 140–57, 2003.
46. Damodaran S. Protein denaturation. In: Hi YH (ed.). Handbook of food science, technology and engineering, CRC Press, FL, 2006. CrossRef
47. Rodrigo SRL, Eleni G, Roberto DS. Characterization and composition of thermostability of purified beta-glucosidase from mesophilic Aureobasidium pullulans and a thermophilic Thermoascus aurantiacus. Process Biochem 2007;42:1101–6. CrossRef