1. INTRODUCTION
Chitin, the second most abundant polysaccharide in nature after cellulose, is a linear polymer of β-1, 4-N-acetylglucosamine. Bacteria, fungi, yeasts, actinomycetes, and plants are the main sources of chitin [1] Complete hydrolysis of chitin can be carried out by a group of chitinolytic enzymes including exochitinase, endochitinase, and chitobiase and release N-acetyl-d-glucosamine subunits [2]. Chitinases have a wide range of applications such as biocontrol agents and biopesticides [3]. They were considered as the best biological control agents of plant pathogens due to their ability to degrade fungal cell walls [4,5]. Trichoderma species were reported to be act as mycoparasites against soil-borne pathogens such as Rhizoctonia solani, Sclerotinia rolfsii, and Fusarium sp. [6-8]. In the present study, the optimal culture conditions for maximum production of chitinase production by Trichoderma strains under solid-state fermentation were studied and results were discussed.
2. MATERIALS AND METHODS
2.1. Isolation of Trichoderma
For this study, Trichoderma strains isolated from rhizosphere soil samples collected from tomato fields of five different areas of Niaye zone, the main area of horticulture production in Senegal: UCAD, Sangalkam, Gorome, Notto Gouye Diama, and Mboro was used. Identification of Trichoderma isolates was based on culture characters as well as microscopic parameters (conidiophores branching, phialides shape and position, spore size, and shape) [9]. The pure cultures maintained at 4°C were used for further studies. The best producer of enzyme was identified by 18S rRNA sequencing (Macrogen, South Korea) as Trichoderma asperellum.
2.2. Screening of Isolates for Chitinase Production
The chitinase activities were determined using chitinase detection medium [10] with slight modifications (colloidal chitin 10 g, MgSO4 0.3 g, NH4SO4 3.00 g, KH2PO4 2.00 g, agar 20.00 g, and distilled water 1000 ml). Colloidal chitin prepared from commercial chitin was used as a carbon source in the above medium. Four days after incubation, a clear zone surrounding the colony indicates the positive chitinase activity. For positives strains, diameter of hyaline zone around the colony (mm) at different incubation periods (days) was measured.
2.3. Chitinase Assay
For the determination of chitinase activity, the reaction mixture containing 1 ml each of crude enzyme and 1% colloidal chitin in 0.05 M phosphate buffer (pH 7.0) was incubated at 35°C for 1 h [2]. After incubation, 1 ml of reaction mixture was taken and to this, 1 ml of distilled water was added. The contents were then boiled in a centrifuge tube for 10 min and centrifuged. An aliquot of 0.5 ml of the supernatant was taken and to this, 0.1 ml of potassium tetraborate was added and boiled for exactly 3 min in a water bath. After cooling, 3 ml of P-Dimethylaminobenzaldehyde reagent was added to this reaction mixture and the absorbance was measured at 585 nm. A blank was maintained without chitin or enzyme. The amount of N-Acetyl D-Glucosamine released in the supernatant was determined using N-Acetyl D-Glucosamine as the standard. One unit of the chitinase activity was defined as the amount of the enzyme producing 1 μ mole of N-Acetate glucosamine per minute in 1 ml of reaction mixture under the standard assay conditions [11].
2.4. Optimization of Parameters for Chitinase Production by T. asperellum
The isolate Trichoderma TG4, highest producer of chitinase activity, was used for optimization using various single parameters. It included incubation period, pH, temperature, carbon sources, nitrogen sources, and colloidal chitin concentration. For each parameter, biomass production (mg/100 ml) and chitinase activity (U/ml) were determined. The effect of incubation period on chitinase production was estimated at different incubation periods (24, 48, 72, 96, and 120 h). Effect of initial pH was studied by adjusting medium pH from 4 to 10 using 0.1 N HCl/NaOH. Six different temperatures (4°C, 15, 20°C, 25°C, 30°C, and 35°C), five carbon sources (sucrose, maltose, galactose, glucose, and colloidal chitin) and five nitrogen sources (NaNO3, (NH4)SO4, yeast extract, beef extract, and peptone) and five chitin concentrations (0.1, 0.5, 1.0, 1.5, and 2.0) were also evaluated. For each parameter optimization, three sets of independent experiments were carried out and the average values are reported.
2.5. Statistical Analysis
Three replicates were maintained for each treatment. Values were given as means ± standard deviation for triplicate samples.
3. RESULTS AND DISCUSSION
3.1. Chitinase Screening
Five of the 20 strains screened showed the positive results for chitinase activity by the formation of clear zone surrounding the colony on chitin agar plates after 72 h of incubation [Table 1]. Maximum diameter of hyaline zone around colony was observed after 5 days of incubation with T. asperellum TG4 (16 mm).
3.2. Effect of Incubation Period on Chitinase Production
Our results [Table 2] elucidated that the incubation period influences the enzyme activity and biomass production. This results showed that a progressive increase production of biomass and enzyme activity by T. asperellum was observed between 3 and 5 days of incubation and a drastic reduction of the two parameters 6 and 8 days. Maximum biomass production (353.3 mg/100 ml) and chitinase activity (2.81 U/ml) were reached at 5 of days of incubation.
3.3. Effect of pH on Chitinase Production
The biomass production and enzyme activity were estimated between pH 3 and 10 [Table 3]. The pH of the medium showed a significant influence on the biomass production and chitinase activity by TG4 Trichoderma strains. The maximum biomass production (453.3 mg/100 ml) and enzyme activity (2.81 U/ml) were observed at pH 6. Our results showed a progressive increase of enzyme activity from pH 4 to 6. In basic pH (8–10), a drastic reduction of biomass production and enzyme activity was observed.
3.4. Effect of Temperature on Chitinase Production
For the effect of different temperature on biomass and chitinase production by TG4 Trichoderma strains, our investigations showed that the maximum biomass production (378.2 mg/100 ml) and enzyme activity (2.99 U/ml) were obtained at 30°C followed by 25°C [Table 4]. Biomass and chitinase activity are low at 4° and increased at 15°C, 20°C, and 25°C. At 35°C and 40°C, a reduction of the different parameters was observed.
3.5. Effect of Carbon Source on Chitinase Production
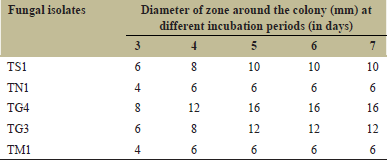 | Table 1: Screening for chitinase production by fungi isolated from Senegal. [Click here to view] |
 | Table 2: Effect of incubation period on chitinase production by Trichoderma asperellum. [Click here to view] |
 | Table 3: Effect of pH on chitinase activity by Trichoderma asperellum. [Click here to view] |
All the carbon sources (sucrose, maltose, galactose, glucose, and colloidal chitin) increase the biomass production and enzyme activity compares to control [Table 5]. Addition of colloidal chitin in the medium gave maximum biomass production (553.3 mg/100 ml) and chitinase activity (2.01 U/ml) followed by sucrose (1.76 U/ml for enzyme activity and 411.6 mg/100 ml for biomass production). The galactose showed the lowest chitinase production (0.94 U/ml) compared to control.
3.6. Effect of Colloidal Chitin Concentration on Chitinase Production
Different concentrations of colloidal chitin (0.1%, 0.5%, 1.0%, 1.5%, and 2.0%) were selected to study the effect on biomass and chitinase production [Table 6]. Our results showed a gradual increase of the two parameters with the increase of colloidal chitin concentration up to 1% that showed the maximum biomass production (353.3 mg/100 ml) and enzyme activity (2.81 U/ml). After that, a reduction of biomass and chitinase activity was observed.
3.7. Effect of Nitrogen Sources on Chitinase Production
Effect of nitrogen sources on IAA production by Trichoderma strains was studied by the addition of various nitrogenous compounds (NaNO3, (NH4)2SO4, yeast extract, beef extract, and peptone). These different nitrogen sources have a significant effect on biomass production and enzyme activity compare to control [Table 7]. Among all the nitrogen sources used, yeast extract was found to be the best nitrogen source for biomass production (456.6 mg/100 ml) and enzyme activity (2.89 U/ml). It is followed for enzyme activity, respectively, by peptone (2.72 U/ml) and beef extract (2.35). Between the nitrogen sources used, (NH4)2 SO4 showed the lowest biomass production (180 mg/100 ml) and chitinase activity (1.04 U/ml).
Several Trichoderma species are able to produce many enzymes like chitinase involved on biocontrol of many soil-borne pathogenic fungi. However, the production of these enzymes depends on many factors such as incubation periods, pH, temperature, carbon sources, and nitrogen sources. Among the 20 strains screened, the results of this study showed that the strains TG4 identify by 18S rRNA sequencing T. asperellum showed the maximum zone of inhibition after 5 days of incubation. Optimization of chitinolytic activity of TG4 showed a maximum chitinase and biomass production at 5 days of incubation at 30°C, pH 6. The decrease of chitinase yield after the optimum period of incubation was probably due to the reduced nutrient level of the medium affecting the enzyme synthesis by the fungus [12,13]. Variation of chitinase production of pH and temperature was observed by many authors. Similarly, to our results Mallikharjuna Rao et al. [14], Ulhoa and Peberdy [15] found maximum chitinase production at 30°C and pH 6. Sandhya et al. [13] found maximum chitinase production after 96 h of incubation at pH 4 using Trichoderma harzianum in submerged fermentation. Nampoothiri et al. [12] also observed maximum chitinase after 96 h of incubation at 30°C, pH 4.5 with T. harzianum in solid-state fermentation. Aida et al. [16] showed high level of chitinase production in culture medium with pH 5 at 30°C for 5 days at shaking condition using Aspergillus terreus species. Increase in chitinase production at acidic pH in the present study was in coincidence with the results of El-Katatny et al. [17], in T. harzianum Rifai. Among the various carbon sources tested, colloidal chitin showed the maximum biomass and chitinase production. Same results were observed by Sandhya et al. [13] and Nampoothiri et al. [12]. Rao et al. [16] also showed that maximum chitinase production was observed when chitin or dried cell walls of S. rolfsii were incorporated in the media. The production of extracellular enzymes like chitinase increases significantly when Trichoderma spp. are grown in media supplemented with either autoclaved mycelium or isolated purified host fungal cell walls [18,19]. Our results showed that yeast extract was the best nitrogen source for biomass and chitinase production by Trichoderma strains. Similar results was reported by Nampoothiri et al. [12], while Sandhya et al. [13] reported that peptone and tryptone showed maximum enzyme production by T. harzianum.
 | Table 4: Effect of temperature on chitinase activity by Trichoderma asperellum. [Click here to view] |
 | Table 5: Effect of carbon sources on chitinase activity by Trichoderma asperellum. [Click here to view] |
 | Table 6: Effect of colloidal chitin concentration by Trichoderma asperellum. [Click here to view] |
 | Table 7: Effect of nitrogen sources on chitinase activity by Trichoderma zasperellum. [Click here to view] |
4. SUMMARY
The present study reports the factors affecting chitinase production by Trichoderma strains. Of the 20 Trichoderma strains isolated, five showed chitinolytic activity. Among the five strains, TG4 (T. asperellum) showed maximum chitinase activity. Optimization studies on T. asperellum showed maximum chitinase activity at 5 days of incubation, pH 6.0, and 30°C temperature. Addition of colloidal chitin and yeast extract as carbon and nitrogen sources, respectively, greatly influenced the chitinolytic activity of Trichoderma strains. These observations, thus, confirmed that T. asperellum (TG4) could be a potential fungus for the production of extracellular chitinase.
5. ACKNOWLEDGMENTS
The authors are thankful to CV Raman International Fellowship for African Researcher of the Department of Science and Technology, Government of India, for the financial assistance under Fellowship Project (DO NO. DST/INT/CVRF/2016). We are also thankful to the Department of Botany and Microbiology, Acharya Nagarjuna University, for providing the facilities to do this work.
6. REFERENCES
1. Pandey S, Shahid M, Srivastava M, Sharma A, Singh A, Kumar V, et al. Chitinolytic assay for Trichoderma species isolated from different geographical locations of Uttar Pradesh. Afr J Biotechnol 2014;13:4246-50. CrossRef
2. Vyas P, Deshpande MV. Chitinase production by Myrothecium verrucaria and its significance for fungal mycelia degradation. J Gen Appl Microbiol 1989;35:343-50. CrossRef
3. Patil RS, Ghormade VV, Deshpande MV. Chitinolytic enzymes: An exploration. Enzyme Microb Technol 2000;26:473-83. CrossRef
4. Hoster F, Schmitz JE, Daniel R. Enrichment of chitinolytic microorganisms: Isolation and characterization of a chitinase exhibiting antifungal activity against phytopathogenic fungi from a novel streptomyces strain. Appl Microbiol Biotechnol 2005;66:434-42. CrossRef
5. Solanki MK, Singh N, Singh RK, Singh P, Srivastava AK, Kumar S, et al. Plant defense activation and management of tomato root rot by a chitin-fortified Trichoderma/Hypocrea formulation. Phytoparasitica 2011;39:471. CrossRef
6. Harman GE, Hayes CK, Lorito M, Broadway RM, Di Pietro A, Peterbauer C, et al. Chitinolytic enzymes of Trichoderma harzianum: Purification of chitobiosidase and endochitinase. Phytopathology 1993;83:313-8. CrossRef
7. Haran S, Schickler H, Oppenheim A, Chet I. Differential expression of Trichoderma harzianum chitinases during mycoparasitism. Phytopathology 1996;86:980-5. CrossRef
8. Madi L, Katan T, Katan J, Henis Y. Biological control of Sclerotium rolfsii and Verticillium dahliae by Talaromyces flavus is mediated by different mechanisms. Phytopathology 1997;87:1054-60. CrossRef
9. Nagamani A, Kunwar IK, Manoharachary C. Handbook of Soil Fungi. New Delhi: IK international; 2006. p. 496p.
10. Agrawal T, Kotasthane AS. Chitinolytic assay of indigenous Trichoderma isolates collected from different geographical locations of Chhattisgarh in Central India. Springerplus 2012;1:73. CrossRef
11. Mathivanan N, Kabilan V, Murugesan K. Purification, characterization, and antifungal activity of chitinase from Fusarium chlamydosporum, a mycoparasite to groundnut rust, Puccinia arachidis. Can J Microbiol 1998;44:646-51. CrossRef
12. Nampoothiri KM, Baiju TV, Sandhya C, Sabu A, Szakacs G, Pandey A. Process optimization for antifungal chitinase production by Trichoderma harzianum. Process Biochem 2004;39:1583-90. CrossRef
13. Sandhya C, Adapa LK, Nampoothiri KM, Binod P, Szakacs G, Pandey A, et al. Extracellular chitinase production by Trichoderma harzianum in submerged fermentation. J Basic Microbiol 2004;44:49-58. CrossRef
14. Mallikharjuna Rao KL, Siva Raju K, Ravisankar H. Cultural conditions on the production of extracellular enzymes by Trichoderma isolates from tobacco rhizosphere. Braz J Microbiol 2016;47:25-32. CrossRef
15. Ulhoa CJ, Peberdy JF. Regulation of chitinase synthesis in Trichoderma harzianum. J Gen Microbiol 1991;137:2163-9. CrossRef
16. Aida FM, Al-Nusarie S, Taghreed S. Production, optimization, characterization and antifungal activity of chitinase produced by Aspergillus terrus. Afr J Biotechnol 2014;13:1567-78. CrossRef
17. El-Katatny MH, Somitsch W, Robra KH, El-Katatny MS, Gübitz GM. Production of chitinase and β-1, 3-glucanase by Trichoderma harzianum for control of the phytopathogenic fungus Sclerotium rolfsii. Food Technol Biotechnol 2000;38:173-80.
18. Qualhato TF, Lopes FA, Steindorff AS, Brandão RS, Jesuino RS, Ulhoa CJ, et al. Mycoparasitism studies of Trichoderma species against three phytopathogenic fungi: Evaluation of antagonism and hydrolytic enzyme production. Biotechnol Lett 2013;35:1461-8. CrossRef
19. Roy A, Hazra S, Pan S. Irradiation to induce better strains of Trichoderma virens. Indian Phytopathol 2005;58:106-10.