1. INTRODUCTION
The global health and socioeconomic burden imposed by the mosquitoes is well established, with an estimated 219 million malaria cases reported worldwide in 2017 (WHO Report, 2017). In addition to malaria, other vector-borne diseases are responsible and pose a significant threat to humanity, especially in tropical and subtropical countries [1]. The African continent contributes maximum to the total global malaria burden because of the presence of the Anopheles gambiae complex among different Anopheles vector, with over 400 million people living under the risk of infection. The use of chemical insecticides for decades has led to the development of resistance in the vector, making malaria control a challenging task, especially for the highly populated countries like India [2]. Also, the unavailability of an effective vaccine has pushed scientists worldwide to look for novel mechanisms for malaria control [3]. In recent years, transmission-blocking interventions (TBI) have emerged as a potential approach to replace our chemical toolkit and enhance our malaria control capabilities. The TBI has an additional advantage over conventional control methods because it avoids the selective resistance pressure within mosquitoes, as TBI relies on their survival instead of mortality [4].
A variety of proteins from Plasmodium falciparum has been previously tested for transmission blocking [5]. However, recent discoveries suggesting the use of multiple mosquito midgut molecules by P. falciparum has diverted the attention of the scientific community toward enzymes such as aminopeptidase N (APN) [5].
APNs are membrane-bound ubiquitous exopeptidase targeting single amino acids at N-terminal site of the polypeptide chain and have been identified in the midgut epithelial cells of various insect species [6]. Their role as cry toxin-binding proteins in insects was established [7,8] and in previous studies shown that till date more than 20 types of APN were reported in insects [9]. The alanyl aminopeptidase N is the leading malarial TBV immunogen [10]. A midgut specific protein, reported to play an important role in ookinete invasion of Plasmodium in the A. gambiae. Previous studies have shown that APN1 acts as a receptor. It is a glycosyl-phosphatidylinositol-anchored protein for attachment of Plasmodium parasite and later helps in sexual growth [11–13].
Rosenfeld and Vanderberg [6] validated the potential use of APN for malaria transmission blocking in A. gambiae by recording a significantly reduced number of oocysts using rabbit polyclonal antibodies against APN in infected mice [14]. Identified potent epitope from AgAPN1 is highly immunogenic. Similar results were observed in A. stephensi [15], suggesting possible multispecies blocking strategies. Also, antibodies against APN1 were responsible for 73% blockage in A. gambiae and 67% in A. stephensi [13]. Based on the crystal structure of the near-full length APN1, a structure-guided construct has been expressed in Escherichia coli and revealed B cells epitopes as transmission-blocking antigens [16]. Studies on the APN1 have also been identified and characterized from A. stephensi as a candidate for Transmission Blocking Vaccine [17]. Also, by applying omics technologies to understand mosquito biology and evolution, these help to explore the diversity and the variations among mosquitoes [18]. To identify protein-coding genes of A. gambiae based on its genomic sequence, a deep proteomic analysis using high-resolution Fourier transform mass spectrometry for both precursor and fragment ions was carried out [19]. Two isoforms of aminopeptidase N was also identified in Aedes aegypti larval midgut [20]. Five APN genes APN1 [13] and APN2, APN3, APN4, and APN5 [21] have been characterized in A. gambiae, but there is no understanding of molecular evolution in-paralogs. The availability of whole genome sequence of A. gambiae provides the opportunity to fill the gap in knowledge. Therefore, the current study was designed to analyze the organized structure of the paralogous gene structure in A. gambiae.
2. MATERIALS AND METHODS
2.1 Retrieval and analysis of A. gambiae APN paralogous gene sequences
The genomic DNA sequences, cDNA sequences, and amino acid sequences of various paralogous APN were obtained from Vectorbase database (https://www.vectorbase.org) and NCBI for comparison between these paralogous. The functional annotation of genes, number of nucleotides, gene products, orientation of gene and its position on the chromosome and amino acids sequences were analyzed in all paralogous. Distribution of intron and exon and intron phase patterns was obtained by online Gene Structure Display Server [22].
2.2. Conserved domains identification for APN proteins
The conserved domains were identified in all paralogous APN sequences by various domain search tools like NCBI CDD(Conserved domain database) [23], Pfam [24], and InterProScan [25] software, which in turn by searching significantly close orthologous family members. The predicted cleavage sites signal peptides were analyzed by using Signal IP algorithm. Glycosylation and phosphorylation sites were predicted by using the Prosite tool [26]. Possible transmembrane domains in APN proteins were also found by using the TMHMM [27]. The Molecular Weight (MW) and Isoelectric point (pI) were calculated by Compute pI/MW [28]. Multiple sequence alignments were performed with BIOEDIT program with all paralogous sequences. Generation of sequence logo was done by Skylign tool to explore conservation of the sequence of motifs in APN paralogs of A. gambiae.
2.3. Protein–protein interaction (PPI) Network construction
The PPIs for 18 APNs protein were examined from STRING v10.5database [29]. PPIs networks were generated based on some interaction sources, i.e., experiments, text mining, co-occurrence, co-expression, databases, gene fusion, and neighborhood. PPIs that have at least a confidence score between medium ranges of 0.4–0.7 were selected for generation of networks. Topological parameters are also evaluated, i.e., the number of edges, number of nodes, and average node degree, etc. Network visualization was also carried out by Cytoscape v 3.5,1 [30]. Cyto-Hubba [31], a java plugin for Cytoscape software, was used to construct the APN hub of mined PPI network.
2.4. Phylogenetic analysis
Eighteen APN gene sequences retrieved from A. gambiae were compared and analyzed for conserved sites using MEGA6.06 [32]. Generation of the phylogenetic tree was done by using Bootstrap consensus neighbor-joining method for APN paralogous gene sequences.
2.5. Expression profile of the A. gambiae APN
The expression patterns were determined separately for all paralogous genes from MozAtlas (http://mozatlas.gen.cam.ac.uk) [33]. The specific expressions of the individual paralogous genes were analyzed in different tissues (midgut, salivary gland, fat body, ovary, and hemolymph, etc) of A. gambiae.
3. RESULTS AND DISCUSSION
3.1 Comparison of paralogous APN genes in A. gambiae
A total of 18 A. gambiae APN gene sequences were retrieved from the PEST genome on Vectorbase [34] (Table 1). Furthermore, the A. gambiae APN cDNA and protein sequences were searched with NCBI. Data consist of various types of APN, CDS length, protein length, orientation, and chromosomal location, and the number of exons for APN genes was extracted out for further analysis. Each of the 18 paralogous APN genes contains a variable number of exons and introns (Table 2). The lengths of the intron regions varied from 0 to 9 among these 18 APN genes (Fig. 1). APN paralog with accession no. AGAP0013001 had the maximum number of introns while AGAP003926 is intronless. The result indicates a large number of APN isomers are present in A. gambiae mosquitoes.
3.2. Genomic localization of APNs
The APNs were distributed over two chromosomes (X and chromosome 2) of A. gambiae. No APN genes are found to be localized on the third chromosome and at telomere positions. Most of these sequences were found in a cluster on the 2R arm of the chromosome (Table 1). The previous studies identified four types of APN, i.e., APN2, APN3, APN4, and APN5 had been found as a gene cluster located on chromosome 2R [10]. APN2, APN3, and APN4 genes along with four other APNs were found organized on a single locus in the forward orientation, suggesting their co-evolution or duplication from a single gene. These clusters may have a similar function or co-expression in the A. gambiae genome. APN1 are present on a different chromosome (2L) and are organized in the reverse orientation (Table 1). Also, most of the APN genes were located in forward orientation over the 2R arm of the chromosome. The chromosomal location and orientation of APNs are summarized in Figure 1 for easy readability. Clustering of the paralogous genes suggests facilitated functional regulation and have been reported in several organisms [35]. The gene conversion or indirect selective forces are the usual mechanisms for coincidental evolution and clustering of paralogs.
 | Table 1: Detail of paralogous APN genes of A. gambiae retrieved from the Vectorbase and NCBI database. The table covers genomic organization, vectorbase ID, gene orientation, chromosomal location, type of APN, number of nucleotides, exon- intron, ORF region, number of amino acids, Molecular weight, transmembrane domain, isoelectric point and signal peptide. [Click here to view] |
 | Table 2: Structure of APN genes from A. gambiae species. E and I with number represents corresponding exon and intron numbers of the gene. 5ʹ and 3ʹ UTR regions are excluded. [Click here to view] |
 | Figure 1: Representation of APN and their paralogous on X, 2L, and 2R arm of chromosomes. All APNs represented by their geneId and the arrow below this Id indicate forward and reverse orientations of APN gene. Integers at the right side of each bar locate arm name. The scale at the top is in megabases (Mb) and at the end of each arm size in Mb is also mentioned. [Click here to view] |
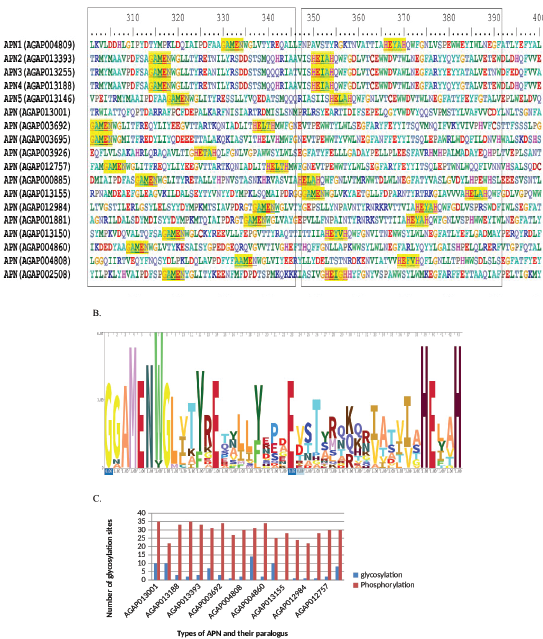 | Figure 2: (A) Amino acid sequence alignment of various paralogous AgAPNs. The conserved gluzincin APN motif represents in box from 320 to 340 aa residues. The zinc-binding conserved sites are shown in box from 335 to 375 aa residues. (B) The APNs HMM logo based on alignment of all paralogous APNs. (C) Showing number of glycosylation and phosphorylation sites in different types of APNs. [Click here to view] |
3.3. Conserved domain identification
Protein BLAST and MSA was performed using all these APN sequences for the known conserved regions. APN proteins characterized by the presence of three domains, aminopeptidase N like, N terminal (IPR042097), Pfam (PF01433) M1 Peptidase (IPR014782), and Pfam (11838) ERAP1-like C-terminal domain (IPR024571) are found in all APNs which is in accordance with the previous studies made in insects and other organisms [36,37]. These domains showed homology to Homo sapiens endoplasmic reticulum aminopeptidase 1 isoform b precursor and human leukotriene A4 hydrolase (PDB Id 2R59). CBB, Pfam, and InterPro also carried out for confirmation of results. GAMEN (300-350 aa), HEXXH (350-390 aa), and WWDVAWLNEGFA (360-400 aa) are highly conserved and they may act as catalytic and zinc-binding sites for peptides (Fig. 2A). The amino acid residues involved in the APN catalytic site of human APN include the amino acid motif GAMEN (residues 352-356) of the human APN [38]. This motif was completely conserved in Anopheles paralogous except for AGAP004808, which have AAMEN. The two H residues in HEXXH act as zinc ion binding sites and E residue is the catalytic site found in all APNs. APNs molecular weight varied from 97 kDa to 220 kDa. There was a slightly low variation in the PI values, ranging from 4.5 to 5.7. Sequence logo was generated to explore the sequence conservation of sequences of APN motifs in A. gambiae (Fig. 2B). Analysis of the predicted phosphorylation and glycosylation sites of APN sequences carried out by using the PROSITE program. It shows a maximum of 14 glycosylation are present in AGAP002508 (Fig. 2C). The previous studies show that three glycosylation sites are present in APN2, APN3, and APN4 [21] and one in APN1 [16]. Analysis of phosphorylation sites in these sequences shows that the number lies between 20 and 35. AGAP013001 has the maximum number, and APN5 is found to have the minimum number of phosphorylation sites.
 | Figure 3: Protein–protein interaction network analysis. Each node represents proteins and edges depict PPI in the network. (A) PPI network of five types of APNs (APN 1 (1278415), AGAP000885 (1277402), AGAP013001 (11176075), AGAP003695 (1274366), and AGAP002508 (1273453). (B) Representation of these five APN hub proteins. (C) PPI network of APN4 (11175889). (D) Rest all types of APN (center circle) shows this type of network. [Click here to view] |
3.4. Protein-protein interaction (PPI)
By using the STRING database, we mined the PPIs for 18 APNs protein based on the experiment, protein homolog, database evidence, text mining, co-expression, and co-occurrence available at STRING database. The search includes all these parameters to explore the PPI data thoroughly. The extracted analysis ranged from average confidence scores (CI-0.4 ≤ S ≤0.7). PPI data revealed that five APNs, APN1, AGAP000885, AGAP013001, AGAP003695, and AGAP002508 possessed more than four interacting partners in the network and these APNs were associated with each other (Fig. 3A). This group of proteins comprising nine more proteins and were found to having a strong connection with each member having 10–11 interacting partner. Also, enrichment and topological information of APNs system input were also analyzed by STRING. The statistics of the network revealed that this intercome comprised of 55 edges and 11 nodes. The average local clustering coefficient and node degree of the network were found to be 0.982 and 9.82. The majority of these A. gambiae system proteins were significantly enriched mainly in the biological process associated with glutathione synthase protein involved in glutathione metabolism. In Cytoscape plugin, cytohubba was used to explore the hub among above said five APNs from STRING TA system PPI network (Fig. 3B). PPI network determined some highly connected nodes that correspond with specific biological functions called Hub proteins. These hub proteins show evolutionary conservation than non-hubs. So, removal of one of protein from this hub may lead to disruption of the network, and thus this can be considered as attractive blocking targets to reduce malaria. APN4 (AGAP013188) shows 11 nodes and 19 edges associated with cadherin protein (experimentally determined) and also co-expressed with alkaline phosphatises (Fig. 3C). The average local clustering coefficient and node degree of the network were found to be 0.855 and 3.45. On the other hand, remaining 12 types of APNs are connected separately with different types of interacting partners, i.e., alkaline phosphatase by 10 node and 11 edges, these are not showing any type of network between them (Fig. 3D). In Lepidoptera, APNs, along with alkaline phosphatases, play a major role in inserting Cry1A toxin oligomers into the cell membrane [21]. In A. aegypti, it is reported that alkaline phosphatase acts as a functional receptor for Cry 11Aa toxin [39]. Anopheles gambiae APN2 also reported to have a receptor for Cry 11Ba [40,41]. So, PPIs network with APN2 and other APNs with alkaline phosphatase play a role in Cry toxin binding.
 | Figure 4: APN Metabolic pathway produced by KEGG, red color denotes the site where APN (APN1, AGAP000885, AGAP013001, AGAP003695, and AGAP002508) proteins are utilized to carry out glutathione metabolism. [Click here to view] |
3.5. Targeting and pathway study
This study showed that different types of APN proteins contained 900 amino acids to 1,060, at the N-terminal region, where 22–25 aa acts as a signal peptide. However, two types of APNs are without signal peptide (Table 1). Experimental evidence shows that most of the APNs were localized to the microvilli in the posterior midgut. Subcellular localization prediction suggests that all APNs were localized in the plasma membrane, so signal peptide is required for Membrane trafficking from Endoplasmic reticulum (ER) to Golgi transport, a forward secretory pathway in the form of Exosomal proteins of microglial cells. KEGG results show that five types of APN proteins including APN1, AGAP000885, AGAP013001, AGAP003695, and AGAP002508 act as a hub connected to glutathione synthase (1271707 in Fig. 3) may involve together in metabolic pathways for metabolism of Glutathione (Fig. 4). APNs of mosquitoes are attached to the midgut surface where they are preferentially localized in cholesterol-rich membrane rafts. Recent evidence suggests that lipid rafts may be an essential component of ookinete invasion of the midgut epithelium. Mosquito-based TBI antigens Anopheles alanyl aminopeptidase N (AnAPN1) [13] known ookinete-interacting proteins were found associated with apical midgut-microvilli detergent-resistant membranes (DRM), which are enriched in lipid rafts, it is reported that other paralogous APNs AGAP003695, AGAP003926, AGAP013146, AGAP012757, AGAP002692, AGAP001881, AGAP013393, AGAP013188, AGAP013255, and AGAP04860 were also present in DRM [42]. APNs of Lepidoptera are anchored to the midgut surface where they are preferentially confined in cholesterol-rich membrane rafts. A complex set of events that require lipid raft integrity, lateral mobility, and partitioning on midgut surfaces has been shown to be essential for Cry1a toxin insertion and pore formation in Heliothis virescens and Manduca sexta [43].
3.6. Expression profiles of APNs at various tissue level
Data are available at MozAtlas (http://mozatlas.gen.cam.ac.uk) on the expression of paralogous APN genes concerning different target tissues, i.e., midgut, malpighian tubules, head, testis, ovary, carcass, and salivary glands. These are collected from EST library and it shows that different types of APN genes were expressed in various tissues [33]. AGAP001881 and AGAP013155 only expressed in midgut tissue. From APN1 to APN5, AGAP012745, AGAP003692, and AGAP012757 showed their highest expression level in the midgut with some expression in the malpighian tubules (Table 3). However, AGAP003695 showed lower expression in the midgut and much higher expression in the malpighian tubules. APN5 showed expression in the midgut, carcass, and the malpighian tubules with very small expression in the head, testis, and ovary. In fact, AGAP013001 shows their expression in carcass and head. AGAP000885 expressed in carcass only. AGAP004808 shows most of their expression in the testis and less expression in the midgut. However, AGAP002508 is expressed in the midgut and the ovary. Also, the expression level of AGAP04860 was observed in the midgut, malpighian tubules, and salivary gland with less expression in the head and the testis. Among all these APNs, most of their expressions were seen in the midgut. It was interesting to note that most of APNs were not expressed in the salivary gland, head, ovarian and testis tissue.
3.7. Phylogenetic analysis
The phylogenetic construction revealed that each branch of tree evolves independently from the other, showing different paralogous APN gene sequences (Fig. 5). Phylogenetic analysis of A. gambiae genome showed a distinct organization of APN gene family. The phylogenetic tree opens two major clusters, including 15 APNs, which are in one group and three APNs in the second cluster. A clade formed by APN2, APN3, and APN4 indicates that these genes are closely evolved [21, 41 and 45]. APN1 is out of the above-said clade and showed that the evolution of these genes occurs independently from their common ancestors. However, APN3 acts as a sister-group to APN2 and APN4, with 100% bootstrap support (Fig. 5). APN5 branched off next, clustering with APN4, APN3, and APN2. Among these five APN genes, APN2, APN3, APN4, and APN5 formed separate single clusters revealing their similar evolution as compared to APN1 genes which formed a different group.
 | Table 3: Expression pattern of A. gambiae APN in different tissues based on data obtained from MozAtlas. [Click here to view] |
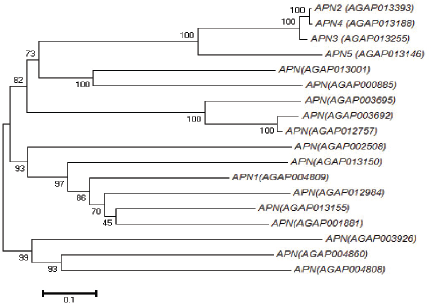 | Figure 5: Showing phylogenetic analysis of different types of APNs with their Accession No. [Click here to view] |
4. CONCLUSION
Through genome-wide analysis of the APN paralogous genes in A. gambiae, covered insilico study on gene structure, location on chromosome, protein properties, functional motifs analysis, protein-protein interaction study, metabolic pathway, and phylogeny. Previous studies are limited to the function of only five types of APNs (APN1-5). Further functional characterization and expression analysis of other APNs will be necessary to get more thoughtful understanding of the evolution of the A. gambiae APN gene families. TBVs promise a more efficient way to malaria control. The previous studies have identified an epitope from A. gambiae APN1 is highly immunogenic. But, there is a weakness due to improper folding of the protein in addition to transmission irrelevant epitopes dilute the production of functional antibodies. In this study, some proteins are identified as hub proteins and may express together during ookinete invasion. So, targeting these hub APNs or other protein from a protein-protein interaction network may also be able to induce more antibody response that significantly inhibits parasite development and may be more effective to control malaria.
ACKNOWLEDGMENTS
The author Renu Jakhar highly acknowledges the financial assistance and support provided by DBT-IPLS, Govt. of India. The authors also would like to thank Arvind Sharma at the University of Nevada, Reno (USA) for valuable comments and suggestions.
AUTHOR’S CONTRIBUTION
Renu Jakhar conducted the study, performed in silico analysis and wrote the manuscript. S.K. Gakhar planned the study. Ritu Gill revised the manuscript.
CONFLICT OF INTERESTS
There is no conflict of interests.
REFERENCES
1. Singh N, Shukla M, Chand G, Barde PV, Singh MP. Vector-borne diseases in central India, with reference to malaria, filaria, dengue and chikungunya. WHO South-East Asia J Public Health 2014;3(1):28.
2. Raghavendra K, Velamuri PS, Verma V, Elamathi N, Barik TK, Bhatt RM et al. Temporo-spatial distribution of insecticide-resistance in Indian malaria vectors in the last quarter-century: need for regular resistance monitoring and management. J Vector Borne Dis 2017;54(2):111.
3. Piermarini P, Esquivel C, Denton J. Malpighian tubules as novel targets for mosquito control. Int J Environ Res Public Health 2017;14(2):111.
4. Wangdi K, Gatton ML, Kelly GC, Banwell C, Dev V, Clements AC. Malaria elimination in India and regional implications. Lancet Infect Dis 2016;16(10):e214–24.
5. Bokharaei H, Raz A, Zakeri S, Djadid ND. 3'-RACE Amplification of aminopeptidase N gene from Anopheles stephensi applicable in transmission blocking vaccines. Avicenna J Med Biotechnol 2012;4(3):131–41.
6. Rosenfeld A, Vanderberg JP. Plasmodium berghei: induction of aminopeptidase in malaria resistant strain of Anopheles gambiae. Exp Parasitol 1999;93(2):101–4.
7. Gill .SS, Cowles. EA, Francis .V. Identification, isolation, and cloning of a Bacillus thuringiensis CryIAc toxin-binding protein from the midgut of the lepidopteran insect Heliothis virescens. J Biol Chem 1995;270:27277–82.
8. Knight .PJ, Knowles. BH, Ellar. DJ. Molecular cloning of an insect aminopeptidase N that serves as a receptor for Bacillus thuringiensis CryIA(c) toxin. J Biol Chem 1995;270:17765–70.
9. Chen, Aimanova, S.Pan, S.S.Gill. Identification and characterization of Aedes aegypti aminopeptidase N as a putative receptor of Bacillus thuringiensis Cry11A toxin. Insect Biochem Mol Biol 2009;39:688–96.
10. Dinglasan RR, Jacobs-Lorena M. Flipping the paradigm on malaria transmission blocking vaccines. Trends Parasitol 2008;24:364–70.
11. Dinglasan RR, Fields I, Shahabuddin M, Azad AF, Sacci JB Jr. Monoclonal antibody MG96 completely blocks Plasmodium yoelii development in Anopheles stephensi. Infect Immun 2003;71(12):6995–7001.
12. Dinglasan RR, Valenzuela JG, Azad AF. Sugar epitopes as potential universal disease transmission blocking targets. Insect Biochem Mol Biol 2005;35(1):1–10.
13. Dinglasan RR, Kalume DE, Kanzok SM, Ghosh AK, Muratova O, Pandey A. Disruption of Plasmodium falciparum development by antibodies against a conserved mosquito Midgut antigen. Proc Natl Acad Sci USA 2007;104(33):13461–6.
14. Mathias DK, Plieskatt JL, Armistead JS, Bethony JM, Abdul-Majid KB, McMillan A, et al. Expression, immunogenicity, histopathology, and potency of a mosquito-based malaria transmission-blocking recombinant vaccine. Infect Immun 2012;80(4):1606–14.
15. Armistead JS, Morlais I, Mathias DK, Jardim JG, Joy J, Fridman A, et al. Antibodies to a single, conserved epitope in Anopheles APN1 inhibit universal transmission of falciparum and vivax malaria. Infect Immun 2013;82(2):818–29.
16. Atkinson SC, Armistead JS, Mathias DK, Sandeu MM, Tao D, Borhani-Dizaji N, et al. Structural analysis of Anopheles midgut aminopeptidase N reveals a novel malaria transmission-blocking vaccine B-cell epitope. Nat Struct Mol Biol 2015;22(7):532.
17. Shabani S, Karimi A, Mahboudi F, Poursharif A, Djadid ND, Motalleb G, et al. Molecular identification and characterization of aminopeptidase N1 from Anopheles stephensi: A candidate for transmission blocking vaccines. Gene Rep 2016;5:157–62.
18. Ruzzante L, Reijnders MJMF, Waterhouse RM. Of genes and genomes: mosquito evolution and diversity. Trends Parasitol 2019;35(1):32–51.
19. Chaerkady R, Kelkar DS, Muthusamy B, Kandasamy K, Dwivedi SB, Sahasrabuddhe NA, et al. A proteogenomic analysis of Anopheles gambiae using high-resolution Fourier transform mass spectrometry. Genome Res 2011;21(11):1872–81.
20. Pootanakit K, Angsuthanasombat C, Panyim S. Identification of two isoforms of aminopeptidase N in Aedes aegypti larval midgut. J BiochemMol Biol 2003;36(5):508–13.
21. Zhang R, Hua G, Andacht TM, Adang MJ. A 106-kDa aminopeptidase is a putative receptor for Bacillus thuringiensis Cry11Ba toxin in the mosquito Anopheles gambiae. Biochemistry 2008;47:11263–72.
22. Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 2015;31(8):1296–7.
23. Marchler BA, Bo Y, Han L, He J, Lanczycki CJ, Lu S, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 2016;45(D1):D200–3.
24. Schaeffer RD, Liao Y, Cheng H, Grishin NV . ECOD: new developments in the evolutionary classification of domains. Nucleic Acid Res 2017;45(D1):D296–302.
25. Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ et al. InterPro in 2017 beyond protein family and domain annotations. Nucleic Acids Res 2016;45(D1):D190–9.
26. Sigrist CJA, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, et al. New and continuing developments at PROSITE. Nucleic Acids Res 2013;41:D344–7; doi: 10.1093/nar/gks1067
27. Gábor E. Tusnády, István S, The HMMTOP transmembrane topology prediction server. Bioinformatics 2001;9(17):849–50.
28. Bjellqvist B, Hughes GJ, Pasquali Ch, Paquet N, Ravier F, Sanchez J, et al. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 1993;14:1023–31.
29. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2014;43(D1):D447–52.
30. Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics 2014;(1):8–13.
31. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;(4):S11.
32. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol 2013;30:2725–9.
33. Baker DA, Nolan T, Fischer B, Pinder A, Crisanti A, Russell S. A comprehensive gene expression atlas of sex-and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics 2011;(1):296.
34. Mongin E, Louis C, Holt RA, Birney E, Collins FH. The Anopheles gambiae genome: an update. Trends Parasitol 2004;20(2):49–52.
35. Deng XJ. Gene expression divergence and evolutionary analysis of the drosomycin gene family in Drosophila melanogaster. J Biomed Biotechnol 2009;Article ID 315423:9.
36. Agrawal N, Malhotra P, Bhatnagar RK. Interaction of gene-cloned and insect cell-expressed aminopeptidase N of Spodoptera litura with insecticidal crystal protein Cry1C. Appl Environ Microbiol 2002;68(9):4583–92.
37. Hughes AL. Evolutionary diversification of aminopeptidase N in Lepidoptera by conserved clade-specific amino acid residues. Mol Phylogenet Evol 2014;76:127–33.
38. Wong AH, Zhou D, Rini JM. The X-ray crystal structure of human aminopeptidase N reveals a novel dimmer and the basis for peptide processing. J Biol Chem 2012;287:36804–13.
39. Fernandez LE, Aimanova KG, Gill SS, Bravo A, Soberon M. A GPI-anchored alkaline phosphatise is a functional midgut receptor of Cry11Aa toxin in Aedes aegypti larvae. Biochem J 2006;394:77–84.
40. Zhang, K. Bayyareddy and M. J.Adang. Anopheles gambiae alkaline phosphatase is a functional receptor of Bacillus thuringiensis jegathesan Cry11Ba toxin. Biochemistry 2009;48:9785–93.
41. Zhang R, Hua G, Urbauer JL, Adang MJ. Synergistic and inhibitory effects of aminopeptidase peptides on Bacillus thuringiensis Cry11Ba toxicity in the mosquito Anopheles gambiae. Biochemistry 2010;49:8512–919.
42. Parish LA, Colquhoun DR, Mohien CU, Lyashkov AE, Graham DR, Dinglasan RR. Ookinete-interacting proteins on the microvillar surface are partitioned into detergent resistant membranes of Anopheles gambiae midguts. J Proteome Res 2011;10(11):5150–62.
43. Zhuang M, Oltean DI, Gómez I, Pullikuth AK, Soberón M, Bravo A, Gill SS. Heliothis virescens and Manduca sextaLipid Rafts Are Involved in Cry1A Toxin Binding to the Midgut Epithelium and Subsequent Pore Formation. J Biol Chem 2002;277(16):13863–72.
44. Hughes AL. Evolutionary diversification of aminopeptidase N in Lepidoptera by conserved clade-specific amino acid residues. Mol Phylogenet Evol 2014;76:127–33.