1. INTRODUCTION
Salvia hispanica L. popularly known as Chia placed under the mint family (Labiateae). This is herbaceous annual plant, native to West central Mexico and Northern Guatemala. During Pre-Columbian times, it was used as food, source of oil and for pharmaceutical purposes by Mesoamericans [1]. Nevertheless, chia has almost disappeared from the markets for 500 years, presumably for religious persecution, and finally due to the difficulties encountered in establishing Chia culture out from America. S. hispanica has been used since ancient times as anticancer, antiulcer, anti-inflammatory [2], cardioprotective and hepatoprotective [3], antidiabetic, antithrombotic, antioxidant, and as antifungal agent. The major phytochemical components reported in S. hispanica are unsaturated fatty acids, myricetin, quercetin, kaempferol and caffeic acid [4]. Recently, renewed interest has emerged for this plant, as researchers have revealed its extraordinary composition and potential beneficial effects on health [5].
Biochemical profiling of chia seeds has shown that they are rich in oil content (32%) of which 60% is α linolenic acid, an omega-3 fatty acid which is allied with various benefits [6]. Unsaturated oil compound has achieved tremendous attention as significant dietary component [7]. Oil extraction from seeds generates a subfraction with high-dietary fiber content (33.9 g/100 g) [8], which contains polyphenols [9], probably associated with antioxidant activity, and also conferring functional qualities to this wonder plant which has various applications in food industry.
Environmental factors, such as change in climate, time, temperature, soil conditions and area of cultivation, play vital roles in the content of bioactive compounds present in S. hispanica L. [10]. Moreover, the quality of seeds is also affected by these factors which are not desirable for commercial production. Tissue culture-based technique provides an adjunct not only to conventional breeding but also for the propagation and genetic improvement of plants. Thus, there is an urgent need for development a highly efficient regeneration protocol for commercial production of S. hispanica L. [11,12].
Tissue culture-induced differentiation creates consistent and competent substitute for mass multiplication to achieve the worldwide demands. Currently, plant tissue culture techniques have been extensively utilized as a means for mass multiplication and germplasm conservation of many plants. The major objective of tissue culture technique is to achieve genetically identical plants to maintain the germplasm, but, during this technique, there is a possibility of genetic variation in regenerated plants known as “somaclonal variations.” Therefore, it is imperative to determine the genetic stability of in vitro regenerated plants. A range of genetic markers such as restriction fragment length polymorphism, random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism, sequence-tagged site, single-nucleotide polymorphism, and inter-simple sequence repeat (ISSR) markers [13,14] are being used for this purpose. In the present study, two DNA markers RAPD [15] and ISSR [16] have been used because of their sensitivity, high reproducibility, reliability, simplicity, and stability to evaluate the genetic diversity of in vitro regenerated plantlets [17-19]. Usages of two DNA-based molecular markers permits improved probability for the identification of genetic variability in the regenerated plantlets as they amplify various regions of the genome. Further, the potential of individual markers to identify genetic variability is not for all time trustable. Therefore, in the current investigation, an effort has been made to develop a rapid, simple, and efficient micropropagation method for regeneration of S. hispanica L. from nodal explants and assessment of genetic stability using RAPD and ISSR markers.
2. MATERIALS AND METHODS
2.1. In Vitro Seed Germination
Certified seeds of S. hispanica L. were used for the present study. Seeds [Figure 1a] were first surface sterilized with 1% (v/v) sodium hypochlorit for 5 min and were washed thoroughly with double-distilled water. The seeds were then inoculated on half-strength MS [20] media containing 3% (w/v) sucrose and 0.8% (w/v) agar which was used as gelling agent. pH of the media was set to 5.8 with 1 N NaOH or 1 N HCl and then autoclaved at 121°C at 15 psi pressure for 15 min. Ten seeds were inoculated in each flask containing about 30 ml of media. Each culture was kept in culture room with 25 μmol photons m−2/s light intensity under 16/8 h of photoperiod and temperature was sustained at 25 ± 2°C. Observations were recorded weekly and were analyzed statistically.
 | Figure 1: Micropropagation of Salvia hispanica. (a) Seed germination, (b) In vitro germinated seedlings after 25 days, (c) shoot buds >2 cm in height used for shoot multiplication experiment, (d) Adventitious shoot production after 4-week culture on a full-strength MS medium supplemented with 3.0 mg/1 6-Benzyladenine, (e) rooting on MS media supplemented with 1.0 mg 1/IBA, (f) regenerated plant in field conditions [Click here to view] |
2.2. Shoot Induction and Multiplication
Nodal explants from 30 days old aseptically germinated seedlings of S. hispanica L. [Figure 1b] were used for tissue culture. Explants were cultured on MS media supplement with 6-benzyladenine (BAP) (1.0–5.0 mg/l) or Kin (1.0–5.0 mg/l) either alone or with α-naphthalene acetic acid (NAA) or indole-3-acetic acid (IAA) (0.1, 0.5, and 1.0 mg/l). Shoot buds induced were subcultured onto fresh media containing BAP (3 mg/l) for multiplication. Observations were taken regularly up to 4 weeks of culture.
2.3. In Vitro Rooting and Field Transfer
The elongated shoots (>3.0 cm in length) with fully expanded leaves were transferred to culture media fortified with IBA, NAA, and IAA (0.5, 1.0, and 2.0 mg/l) for the development of proper root system. Regenerated plantlets with profuse shoot and roots were cautiously removed and rinsed gently with tap water. Rooted plantlets were transferred to plastic cups having garden soil and organic manure (1:1) and kept in growth chamber. To avoid rapid dehydration of in vitro regenerated plants, pots were enclosed with plastic bags to maintain humidity levels and watered with half concentration MS solution every alternate day. Plant growth was monitored for 2 months before transplanting to the open garden soil. The survival rate of regenerated plantlets was calculated.
2.4. Analysis of Genetic Fidelity Using RAPD and ISSR Markers
Genetic fidelity of tissue culture regenerated plants was analyzed using RAPD and ISSR markers. Genomic DNA of aseptically germinated mother plant and 10 in vitro raised plantlets was isolated using Cetyl Trimethyl ammonium bromide assay [21]. Quantitative assessment of genomic DNA was carried out by NanoDrop™ spectrophotometer ND-1000 and 1% (w/v) agarose (HiMedia Laboratories Pvt. Ltd., India) gel electrophoresis, respectively. The final concentration was set to 20 μg/μl for polymerase chain reaction (PCR) and stored at −20°C. 30 RAPD primers and 10 ISSR primers were selected for analyzing the genetic stability of the regenerated plants. RAPD and ISSR reactions were performed in Mycycler™ (Bio-Rad laboratories, India). PCR amplifications were made to 20 μl containing 10 μl master mix, 1.5 μl primer, 1.5 μl of template DNA, and 7 μl autoclaved doubled distilled water. The whole experiment was designed for RAPD reactions had initial denaturation step at 95°C for 3 min; 35 cycles of 30 s at 95°C, primer annealing at 39°C for 30 s, and primer extension at 72°C for 30 s; followed by final extension of 10 min at 72°C. ISSR method was started with an initial denaturation step of 95°C for 3 min; 35 cycles of 30 s at 95°C, primer annealing at 50°C for 30 s, and primer extension at 72°C for 30 s; followed by final extension for 10 min at 72°C. The amplified DNA was separated on 1.5% (w/v) agarose gel and was stained with EtBr (HiMedia Laboratories Pvt. Ltd.) and photographed in a gel documentation system (Bio-Rad laboratories, India). The sizes of the amplification products were calculated with a 1500 bp DNA ladder (HiMedia Laboratories Pvt. Ltd., India). All PCR reactions for RAPD and ISSRs were repeated to confirm their reproducibility, and prominent bands were scored. The data were denoted as “1” for the presence and “0” for the absence of a band. The plant samples which were containing all the bands were represented as monomorphic, whereas those DNA samples that lack some DNA bands were represented as polymorphic.
2.5. Statistical Analysis
The statistical examination of the data revealed from the tissue culture regenerated plants was subjected to analysis of variance, and the significantly different means at the 5% level of significance (P ≤ 0.05) were recognized using Tukey’s HSD test (SPSS software).
3. RESULT AND DISCUSSION
3.1. Shoot Bud Induction and Proliferation
Shoot buds were induced directly from nodal explants on MS medium fortified with different plant growth hormones such as Kin and BAP [Figure 1c]. The highest induction was obtained when the explants were cultured on MS medium fortified with BAP (3.0 mg/l), where an average of 9 shoots buds was induced per explant [Figure 1d]. Increasing or decreasing the concentrations of BAP beyond the optimum level had detrimental effect on in vitro regeneration of shoots buds [Table 1]. Induction of shoot buds was also obtained on media fortified with Kin (3.0 mg/l) where a maximum of two shoot buds per explant was induced which was significantly lower in number than those induced on BAP. Hence, BAP proved to be most efficient cytokinin than Kin for in vitro regeneration of shoots. BAP has also been reported to be the most competent plant growth hormone for induction of multiple shoots in various plants of Salvia including Salvia santolinifolia [22], Salvia brachyodon [23], Salvia africana-lutea [24], Salvia splendens [25], and Salvia officinalis [26]. Contrary to this, Chen et al. [27] found Kin to be more effective for the formation of adventitious shoot buds in Salvia miltiorriza.
The physiological influences of auxins (NAA and IAA) at various doses (0.1, 0.5, and 1.0 mg/l) were investigated with the most favorable cytokinin concentration (BAP; 3 mg/l) in the media for highest plantlet regeneration. Addition of auxin to the culture media did not improve the shoot regeneration efficiency of explants. An average number of shoot buds obtained on MS media containing BAP (3 mg/l) with IAA (0.5 mg/l) was comparable to those obtained on BAP (3 mg/l) alone. Shoot buds induced were sectored and subcultured on fresh media for multiplication of shoot buds on various hormonal combinations after 1 month of culture. Culture medium fortified with BAP (3 mg/l) was also found to be appropriate for proliferation and elongation of shoot buds where an average of 15 shoots were produced per explant. Combination of BAP with auxins has been found to the promontory for regeneration; however, synergistic effect of BAP with auxins was not found optimum for in vitro shoot regeneration in S. hispanica L. and an our results are in accordance with Molina et al. [28] where BAP in combination with auxin was not found suitable for in vitro culture. Similarly, Echeverrigaray et al. [29] and Fraternale et al. [30] reported that the addition of auxin to the regeneration media failed to enhance axillary shoot proliferation. However, Skala and Wysokinska [31] suggested media fortification with BAP and auxins (NAA and IAA) to be more effective for the induction of shoot buds and their subsequent proliferation.
3.2. Root Induction, Acclimatization, and Field Transfer
Regenerated plantlets more than 3 cm in length were cultured on half strength, full strength MS and MS composed with auxins viz. IAA, IBA, and NAA for induction of roots. No rooting response was achieved on half and full strength MS media without any growth regulators. Rooting of regenerated shoots was obtained on medium fortified with various concentrations of auxins - IBA, IAA, and NAA (0.1, 0.5 and 1.0 mg/l) [Table 2]. Highest number of healthy roots was achieved on culture medium containing 1.0 mg l-1 IBA [Figure 1e]. Further increasing the level of IBA decreased the length and number of roots developed. Although roots were induced on NAA and IAA, they were very thin, slender, and were not able to support the plant system. Hence, IBA (1.0 mg/l) proved to be the finest for rooting of the in vitro developed plantlets. The effectiveness of IBA in rooting has been reported in various species of Salvia by Arikat et al. [32] (Salvia fruticosa mill), and Kabir et al. [33] (S. splendens). Maximum rooting on culture medium without growth regulators has been observed in a various plants counting Salvia valentine and Salvia blancoana [34]. However, no rooting response was observed on media devoid of auxins in S. hispanica L. in the current study.
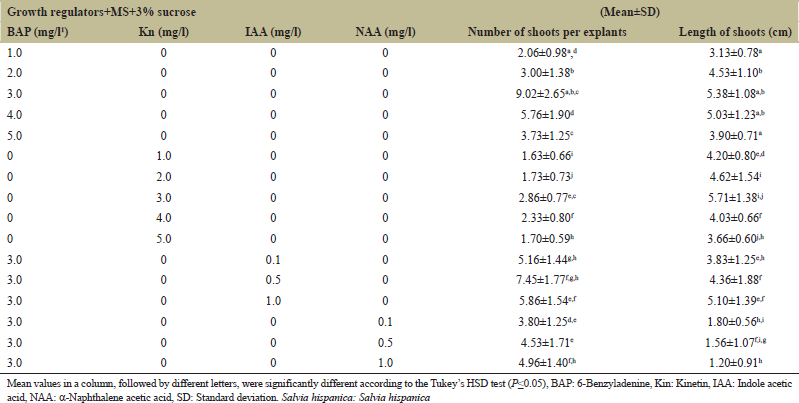 | Table 1: Effect of plant growth regulators on shoot regeneration from nodal explants in S. hispanica L. [Click here to view] |
The rooted plants were acclimatized and hardened before transferring to field conditions. Plantlets with well-developed root system were transferred to plastic cups having garden soil and organic manure (1:1), which were kept in culture room for a period of 15–20 days. Healthy plants were then transferred to earthen pots enclosed with plastic bags to sustain humidity in field conditions [Figure 1f]. The survival rate of field transferred plants was 75%. All plantlets were healthy and showed normal growth.
3.3. Evaluation of Genetic Fidelity of Regenerated Plantlets Using RAPD and ISSR Markers
Genetic uniformity of regenerated shoots was investigated using RAPD and ISSR markers. DNA based markers proved to be useful in differentiating different accessions, species, subspecies, varietal identification, and for genetic mapping of many plants. Molecular markers have been used to identify somaclonal variation in several micropropagated plants as well as genetic diversity between cultivars and germplasm [35-37]. Different types of changes at the genetic level, such as deamplification and amplification of genes, single-nucleotide changes, and alterations in DNA methylation patterns have been related with genetic instability produced under in vitro conditions [38].
A total of 30 arbitrary RAPD primers were used for preliminary studies and among them 12 produced clear and reproducible bands. Total bands which were scorable for each RAPD primer varied from 4 (OPB-09) to 11 (OPA-02). They showed 85 distinct and scorable bands with an average of 7.08 bands per primers [Figure 2a and b]. Every primer has its own a unique set of amplification products ranging in size from 100 bp (OPB-04) to 1300 bp (OPB-02). Details of amplified bands for RAPD primer are presented in Table 3. A total of 819 bands were produced during RAPD analysis and all bands were found to be monomorphic. Primer OPA-02 amplified the maximum band numbers (121), while only 44 bands were produced by primer OPB-09. 10 ISSR primers were used in the preliminary studies of which only five primers formed clear and reproducible bands. They showed 24 distinct and scorable bands in the size range of 250 bp (ISSR-810) to 1200 bp (ISSR-818). Total bands which were scorable for each ISSR primer varied from 3 (ISSR-841) to 6 (ISSR-810), with in an average of 4.8 bands per primer [Table 4]. A total of 261 bands were produced during ISSR analysis and all bands were found to be monomorphic [Figure 2c and d]. Primer ISSR-810, amplified the maximum band numbers (65), while only 33 bands were produced by primers ISSR-841.
Therefore, two DNA-based markers, RAPD, and ISSR were used to access genetic similarity in regenerated plantlets. No genetic variability among regenerates of S. hispanica L. was detected. The results are in accordance with the studies of Saha et al. [39,40] where regenerated plantlets of Ocimum gratissimum, Ocimum basilicum from nodal segments showed genetic stability. Clonal similarity of long-term cultured regenerates of Lavandula officinalis and Thymus persicus was assessed by RAPD markers [41,42]. Samantaray et al. [43] evaluated genetic similarity in in vitro regenerated plantlets of Vitex teifolia by 60 RAPD and 27 ISSR markers. Saha et al. [44] reported an improved and efficient micropropagation of Mentha piperita using nodal explant and assessment of genetic fidelity using RAPD analysis, thereby confirming the genetic uniformity of the in vitro raised plantlets with the donor plant. Al-Rawashdeh [45] studied molecular taxonomy between Ziziphora tenuior, Mentha longifolia, and Mentha spicata populations using the RAPD technique. Some researchers have studied the use of ISSR markers [46-48] shown that ISSR is a simple, rapid, and reproducible means to access genetic diversity among closely related cultivars. Hence, it is an essential to access the genetic stability of in vitro raised plants to develop clonally identical progeny.
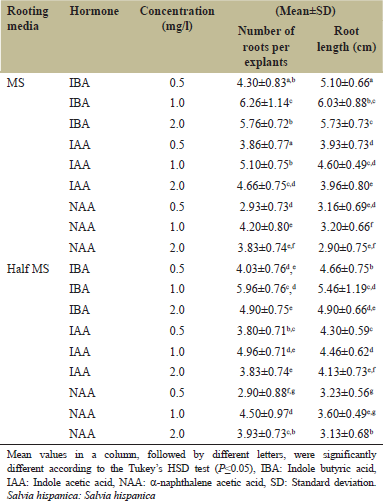 | Table 2: Effect of different auxins IBA, IAA, and NAA on rooting of microshoots in S. hispanica L [Click here to view] |
 | Figure 2: (a) Random amplified polymorphic DNA profiles using the primes OPB-09 (lane M donar plant, lanes 1–10 micro propagited plants, L 1 kb ladder and (b) CPB-10 (lane M donar plant, lanes 1–10 micro propagated plants, L 1 kb ladder, (c) Intersimple sequence repeat (ISSR) profiles using the primers ISSR 836 pane M donar plant, lanes 1–10 micropropagated plants, L1 kb ladder and (d) ISSR 841 pane M donar plant, lanes 1–10 micropropagated plants, 1.1 kb ladder. [Click here to view] |
 | Table 3: List of RAPD primers used to detect polymorphism [Click here to view] |
 | Table 4: List of ISSR primers used to detect polymorphism [Click here to view] |
In our study, genetic similarity of micropropagated plantlets was established using an identical set of RAPD and ISSR markers. However, they did not show any genetic variability among the tissue culture-raised plantlets; hence, nodal explants can be effectively used for the commercial multiplication of S. hispanica L. without much risk of genetic instability.
4. CONCLUSIONS
The present study provides the first report describing a competent in vitro protocol for regeneration of true-to-type plantlets from S. hispanica L. using nodal explant. The protocol describes a rapid, reproducible regeneration method for induction, and proliferation of shoot buds on media composed with BAP (3 mg/l) and maximum induction of roots on MS media with 1.0 mg/l IBA. Furthermore, assessment of genetic fidelity using RAPD and ISSR primers revealed monomorphic bands ascertaining true to the type nature of regenerated plantlets. Therefore, the study has created important information with regard to the efficacy of RAPD and ISSR markers in genetic fidelity studies of in vitro raised plantlets of S. hispanica L., which will be of practical importance in future for screening tissue culture raised plants. The protocol developed in this study provides a rapid multiplication protocol not only for commercial exploitation and can be further used for transformation studies in this plant.
5. ACKNOWLEDGMENT
The authors gratefully acknowledge DRS Phase II, Department of Botany, University of Rajasthan for providing necessary facilities.
6. REFERENCES
1. Jamboonsri W, Phillips TD, Geneve RL, Cahill JP, Hildebrand DF. Extending the range of an ancient crop, Salvia hispanica L.- a new ω3 source. Gen Resour Crop Evol 2012;59:171-8.
2. Simopoulos AP. Omega-3 fatty acids in inflammation and autoimmune diseases. J Am Coll Nutr 2002;21:495-505.
3. Jeong SK, Park HJ, Park BD, Kim IH. Effectiveness of topical chia seed oil on pruritus of end-stage renal disease (ESRD) patients and healthy volunteers. Ann Dermatol 2010;22:143-8.
4. Lu Y, Foo LY. Polyphenolics of Salvia: A review. Phytochemistry 2002;59:117-40.
5. Yadav A, Joshi A, Kothari SL, Kachhwaha S, Purohit S. Medicinal, nutritional and industrial applications of Salvia species: A revisit. Int J Pharm Sci Rev Res 2017;43:27-37.
6. Rosamond WD. Dietary fiber and prevention of cardiovascular disease. J Am Coll Cardiol 2002;39:57-9.
7. Ayerza R. Oil content and fatty acid composition of chia (Salvia hispanica L.) from five northwestern locations in Argentina. J Am Oil Chem Soc 1995;72:1079-81.
8. Craig R, Sons M. Application for approval of whole chia (Salvia hispanicaL.) seed and ground whole chia as novel food ingredients. Advisory Committee for Novel Foods and Processes. Ireland: Company David Armstrong; 2004. p. 1-29.
9. Taga MS, Miller EE, Pratt DE. Chia seeds as a source of natural lipid antioxidants. J Am Oil Chem Soc 1984;61:928-31.
10. Dubois V, Breton S, Linder M, Fanni J, Parmentier M. Fatty acid profiles of 80 vegetable oils with regard to their nutritional potential. Eur J Lipid Sci Technol 2007;109:710-32.
11. Kaneyoshi J, Kobayashi S, Nakamura Y, Shigemoto N, Doi Y. A simple and efficient gene transfer system of trifoliate orange (Poncirus trifoliate Raf.). Plant Cell Rep 1994;13:541-5.
12. Jha TB, Ghosh B. Plant Tissue Culture-Basic and Applied. India: Universities Press; 2005.
13. Sonmezoglu OA, Bozmaz B, Yıldırım A, Kandemir N, Aydın N. Genetic characterization of Turkish bread wheat landraces based on microsatellite markers and morphological characters. Turk J Biol 2012;36:589-97.
14. Phulwaria M, Rai MK, Shekhawat NS. An improved micropropagation of Arnebia hispidissima (Lehm.) DC. and assessment of genetic fidelity of micropropagated plants using DNA-based molecular markers. Appl Biochem Biotechnol 2013;170:1163-73.
15. Williams K, Kubelik AR, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acid Res 1990;18:1631-5.
16. Zietkiewicz E, Rafalski A, Labuda D. Genome fingerprinting by simple sequence repeat (SSR)- anchored polymerase chain reaction amplification. Genomics 1994;20:176-83.
17. Dangi B, Khurana-Kaul V, Kothari SL, Kachhwaha S. Micropropagtion of Terminalia bellerica from nodal explants of mature tree and assessment of genetic fidelity using ISSR and RAPD markers. Physiol Mol Biol Plants 2014;20:509-16.
18. Goyal P, Kachhwaha S, Kothari SL. Micropropagation of Pithecellobium dulce (Roxb.) Benth-a multipurpose leguminous tree and assessment of genetic fidelity of micropropagated plants using molecular markers. Physiol Mol Biol Plants 2012;18:169-76.
19. Ray T, Dutta I, Saha P, Das S, Roy SC. Genetic stability of three economically important micropropagated banana (Musa spp.) cultivars of lower Indo-Gangetic plains, as assessed by RAPD and ISSR markers. Plant Cell Tissue Organ Cult 2006;85:11-21.
20. Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 1962;15:473-97.
21. Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull Bot Soc Am 1987;19:11-5.
22. Tour J, Khalida K. In vitro regeneration of Salvia Santolinifolia. Pak J Bot 2014;46:325-8.
23. Misic D, Grubisic D, Konjevic R. Micropropagation of Salvia brachyodon through nodal explants. Biol Plantarum 2006;50:473-6.
24. Makunga NP, Staden J. An efficient system for the production of clonal plantlets of the medicinally important aromatic plant: Salvia africana-lutea L. Plant Cell Tissue Organ Cult 2008;92:63-72.
25. Xiong LZ, Chun LC, Wei YH, Liang SQ, Yi GW, Ming WH, et al. Micropagation technique of dwarf Salvia splendens. J Fujian Agr Univ 2001;30:483-5.
26. Boldura OM, Radu F, Popescu S, Borozan A. Regeneration, micropropagation, callus cultures and somatic embryogenesis of common sage (Salvia officinalis L.). Bull UASVM Horticult 2010;67:308-13.
27. Chen UC, Shiau YJ, Tsay HS, Hsia CN. Influence of cytokinin and ventilating container closure on shoot proliferation and hyperhydricity of in vitro Salvia miltiorriza culture. J Taiwan Agric Res 2005;54:93-102.
28. Molina MS, Luis A, Luis J. In vitro mass propagation of Salvia canariensis L. by axillary shoots. Acta Soc Bot Polon 1997;66:351-4.
29. Echeverrigaray S, Carrer PR, Andrade LB. Micropropagation of Salvia guaranitica benth. through axillary shoot proliferation. Braz Arch Biol Technol 2010;53:883-8.
30. Fraternale D, Bisio A, Ricci D. Salvia jamensis J. Compton: In vitro regeneration of shoots through TDZ and BA. Plant Biosyst 2013;147:713-6.
31. Skala E, Wysokinska H. In vitro regeneration of Salvia Nemorosa L. from shoot tips and leaf explants. In Vitro Cell Dev Biol Plant 2004;40:596-602.
32. Arikat NA, Jawad FM, Karama NS, Shibli RA. Micropropagation and accumulation of essential oils in wild sage (Salvia fruticosa Mill.). Sci Horticult 2004;100:193-202.
33. Kabir MH, Mamun AN, Roy PK, Islam MR, Jahan MT, Talukder SU. In vitro mass propagation of salvia (Salvia splendens) from Nodal Explant. Nucl Sci Appl 2014;23:51-4.
34. Cuenca S, Amo-Marco JB. In vitro propagation of two Spanish endemic species of Salvia through bud proliferation. In Vitro Cell Dev Biol Plant 2000;36:225-9.
35. Srivastava S, Jain R, Gupta PS, Singh J. Genetic stability of in vitro raised Sugarcane plantlets by RAPD markers Plant. Cell Biotechnol Molecul Biol 2006;7:93-6.
36. Bhowmik SS, Kumaria S, Rao SR, Tandon P. High frequency plantlet regeneration from rhizomatous buds in Mantisia spathulata Schult. and Mantisia wengeri Fischer and analysis of genetic uniformity using RAPD markers. Indian J Ex Biol 2009;47:140-6.
37. Peschke VM, Phillips RL, Gengenbach BG. Genetic and molecular analysis of tissue culture derived Ac elements. Theoret Appl Genet 1991;82:121-12.
38. Rani V, Raina SN. Genetic fidelity of organized meristem-derived micropropagated plants: A critical reappraisal. In Vitro Cell Dev Biol Plant 2000;36:319-30.
39. Saha S, Kader A, Sengupta C, Ghosh PD. In vitro propagation of Ocimum gratissimum L. (Lamiaceae) and its evaluation of genetic fidelity using RAPD marker. Am J Plant Sci 2012;3:64-74.
40. Saha S, Sengupta C, Ghosh P. Evaluation of the genetic fidelity of in vitro propagated Ocimum basilicum L. using RAPD and ISSR markers. J Crop Sci Biotech 2014;17:281-7.
41. Prasad A, Shukla SP, Mathur A, Chanotiya CS, Mathur AK. Genetic fidelity of long-term micropropagated Lavandula officinalis Chaix.: An important aromatic medicinal plant. Plant Cell Tissue Organ Cult 2014;120:1-10.
42. Bakhtiar Z, Mirjalili MH, Sonboli A, Farimani MM, Ayyari M. In vitro propagation, genetic and phytochemical assessment of Thymus persicus-a medicinally important source of pentacyclic triterpenoids. Biologia 2014;69:594-603.
43. Samantaray S, Bishoyi AK, Maiti S. Plant regeneration from callus cultures of Vitex trifolia (Lamiales: Lamiaceae): A potential medicinal plant. Rev Biol Trop 2013;61:1083-94.
44. Saha S, Dey ST, Adhikari S, Ghsoh PD. In vitro multiple shoot regeneration and analysis of genetic fidelity of Mentha piperita L. Bionature 2010;30:71-81.
45. Al-Rawashdeh IM. Genetic variability in a medicinal plant Artemisia judaica using random amplified polymorphic DNA (RAPD) markers. Int J Agric Biol 2011;13:279-82.
46. Joshi P, Dhawan V. Assessment of genetic fidelity of micropropagated Swertia chirayita plantlets by ISSR marker assay. Biol Plant 2007;51:22-6.
47. Xu J, Wang Y, Zhang Y, Chai T. Rapid in vitro multiplication and ex vitro rooting of Malus zumi (Matsumura) Rehd. Acta Physiol Plant 2008;30:129-32.
48. Lata H, Chandra S, Khan I, ElSohly MA. Assessment of genetic stability of micropropagated Cannabis sativa plants by ISSR markers. Planta Med 2010;76:97-100.