1. INTRODUCTION
In the past two decades, advances in molecular biology research have led to extraction, amplification of targeted gene, and consequently, sequencing of DNA from different tissues, blood, non-invasive biological samples including formalin-fixed and ethanol-preserved specimens [1,2]. DNA analysis is a tool for the study of facets of biology, not least of all fields such as evolutionary biology, biosystematics, biodiversity, population biology, conservation biology, and ecology [3]. For the study of population biology and conservation biology, mostly samples were collected from remote areas and preserved in different chemicals [4]. Some chemicals are not appropriate for the DNA studies because they create some modifications in tissue chemicals or degradation of DNA which hinder the molecular biology experiments.
Tissues preservation is most important to make samples keep on original for long time worth for getting a high yield of genomic DNA for molecular studies. Safety of tissue samples for genomic DNA extraction is important as it can protect these potentially to give a high yield of genomic DNA [5]. Usually formaldehyde is most commonly used chemical for preservation of tissues in laboratories. Ethanol solutions are one of the methods for tissue preservation for DNA analysis. Ethanol is appropriate to the storage of vertebrate tissue and has been used successfully in DNA hybridization and sequencing [5,6].
The extraction of the high quality of genomic DNA may be difficult in formalin-fixed tissues as of cross-linking between proteins and DNA as formalin induces DNA destruction and nucleotide modification [7,8]. The rapid reaction of formalin with double helical DNA generally is flexible but over the long term, especially with denaturation of the DNA, a variety of reactions can occur, many of which have not been characterized [8,9].
In the present study, three different preservatives formaldehyde, ethanol 90% solution, and freezing at -40°C were used to examine and determined the effects of DNA of the samples at different time intervals. The aim of this study was to find which method of tissue storage is suitable for extraction of optimum quality of DNA for molecular and phylogenetic studies because the availability of fresh samples in taxonomical studies is limited.
2. MATERIALS AND METHODS
2.1. Collection of Fish Samples
Five specimens of Sperata seenghala were collected from Upper Lake Bhopal brought to the laboratory for performing experimental work. Liver and muscle tissues were taken out for extraction of genomic DNA. Tissues preserved in 8% formalin, 90% alcohol, and -40°C deep freezer for 5 days, 20 months, and 28 months.
2.2. Genomic DNA Isolation and Quantification
Isolation of genomic DNA was performed as the protocol of Janarthan and Vincent [10] using phenol:chloroform:isoamyl-alcohol (25:24:1) method.
Qualitative and quantitative estimation of extracted genomic DNA was done by NanoDrop ultraviolet spectrophotometer (ND-1000) by calculating the ratio of absorbance at 260– 280 nm wavelength. Pure genomic DNA shows ratios as 1.8 at 260 nm v/s 280 nm with respect to protein contamination, because protein tends to absorb at 280 nm wavelength. Final dilutions were made as required for polymerase chain reaction (PCR) amplification ±50 ng/μl of DNA samples.
2.3. PCR Amplification
Two primers RAn-5 and RAn-6 with accession numbers AM750052 and AM765829, respectively, were used for final PCR amplification who gave positive and scorable results. A 25 μl of reaction mixture composed of 12.50 μl Red Dye, 1.0 μl primer, 11.00 μl of sterile distilled water, and 1.0 μl template DNA was prepared for PCR amplification with preheating for 5 min at 94°C. PCR was run for 45 cycles consisted of a 94°C denaturation for 0.45 min, 37°C annealing for 1.0 min, and 72°C elongation for 1.5 min in thermal cycler (Model EP Gradient, Eppendorf, Germany) with a final extension of 72°C for 10 min.
2.4. Gel Electrophoresis and Visualization of DNA Pattern
Amplified DNA fragments were separated by gel electrophoresis on 1.2% agarose gel concentration containing ethidium bromide for visualization of DNA fragments. Low range DNA ladder was also used for each gel with the known interval of 100, 200, 300, 600, 1000, 1500, 2000, 2500, and 3000 bp. Gel was visualized and photographed on gel documentation system (Alpha-Innotech, USA) for scoring of DNA fingerprints and finally obtained molecular weights.
3. RESULTS AND DISCUSSION
Molecular research studies need for genomic DNA extraction from animal tissues including aquatic animals for performing molecular taxonomy, development of species-specific markers, recombinant DNA technology, and transgenic studies. Even few methods exist genomic of DNA extraction from cultured cells and blood, but in the absence of these facilities, how the samples are collected from the field and preserve them in a proper manner so that a desired quality as well quantity of DNA can be obtained from the preserved samples. The major problems challenged by forensic laboratories or researchers engaged in molecular biology are to obtain the samples in proper conditions to performed molecular work. Most of the biological samples brought for DNA analysis are either partially degraded or completely degraded, and many times, it is not possible to extract DNA from them, and therefore, the present research carried on S. seenghala.
In this study, tissues preserved in different preservatives for 5 days gave following results with respect to quantification of DNA. 8% formalin preservative for 5 days, DNA quality was obtained from 8.07 ng/ul to 15.77 ng/ul with absorbance ratio from 1.30 to 1.53 of 260/280 wavelengths [Table 1]. The obtained results were unsatisfactory with respect to quality and quantity as they do not fall in the normal range. In case of -40°C preservative tissue samples, the DNA was recorded from 410.79 ng/ul to 645.79 ng/ul with the ratio between 1.98 and 2.13 of 260/280 [Table 1 and Figures 1-5] showed satisfactory results and may called results are up to the mark, because it lies close to the normal value for obtaining better results in DNA amplification. Laith [11] studied on the effect of formalin, alcohol, and freezing on some body proportion of a marine fish Alepes djeddaba, observed greatest shrinkage of specimens preserved in 8% formalin-tap water, while the least shrinkage was in the fishes stored in 70% alcohol tap water. The present investigation also shows that the tissues preserved in formalin not gave good results.
 | Table 1: Quantification of extracted DNA from tissues of S. seenghala preserved in 90% alcohol, 8% formalin, and – 20°C deep freezer. [Click here to view] |
 | Figure 1: Qualitative and quantitative status of extracted DNA from preserved samples in 90% alcohol for 5 days. [Click here to view] |
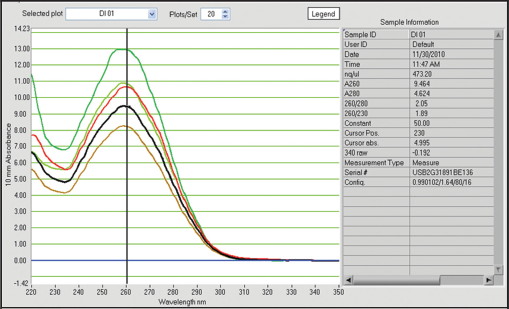 | Figure 2: Qualitative and quantitative status of extracted DNA from preserved samples in 8% formalin for 5 days. [Click here to view] |
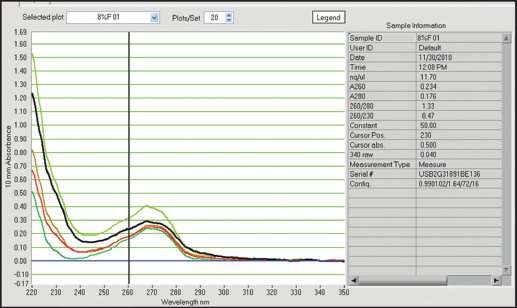 | Figure 3: Qualitative and quantitative status of extracted DNA from preserved samples in dried ice for 5 days. [Click here to view] |
 | Figure 4: Qualitative and quantitative status of extracted DNA from preserved samples in 90% alcohol, 8% formalin for 20 months and fresh samples. [Click here to view] |
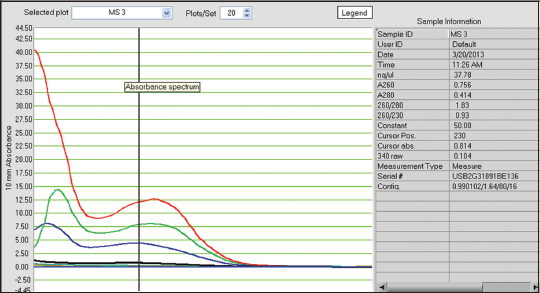 | Figure 5: Qualitative and quantitative status of extracted DNA from preserved samples in 90% alcohol, 8% formalin for 28 months and fresh samples. [Click here to view] |
 | Table 2: Frequency of amplicons and their molecular weight obtained through RAPD marker RAn-5. [Click here to view] |
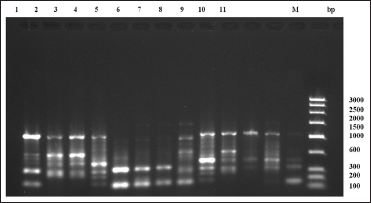 | Figure 6: Random amplified polymorphic DNA-polymerase chain reaction fragment patterns obtained by RAn-05 from tissues of Sperata seenghala preserved for 5 days. 1– 4 = samples with 90% alcohol preservative, 5– 8 = samples with 8% formalin preservative, 9– 11 = samples with dried ice preservative. M is molecular marker (bp) of low range DNA ladder. [Click here to view] |
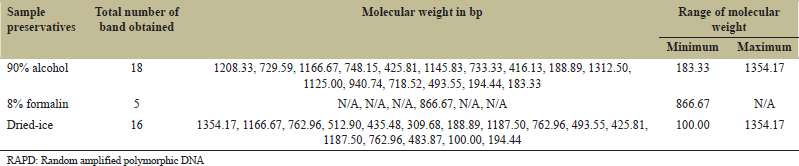 | Table 3: Frequency of amplicons and their molecular weight obtained through RAPD marker RAn.6. [Click here to view] |
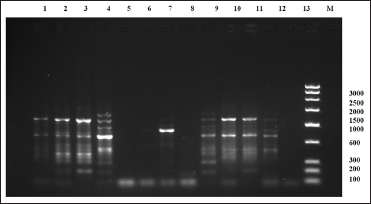 | Figure 7: Random amplified polymorphic DNA-polymerase chain reaction fragment patterns obtained by RAn-06 from tissues of Sperata seenghala preserved for 5 days. 1– 4 = samples with 90% alcohol preservative, 5– 8= samples with 8% formalin preservative, 9– 11= samples with dried ice preservative. M is molecular marker (bp) of low range DNA ladder. [Click here to view] |
  | Figure 8: Random amplified polymorphic DNA-polymerase chain reaction fragment patterns obtained by RAn-05 from tissues of Sperata seenghala preserved for 20 months. 1– 6 = tissues preserved in 8% formalin, 07– 09= tissues preserved in 90% alcohol, 10– 12 tissues preserved in dried ice. M is molecular marker (bp) of low range DNA ladder. [Click here to view] |
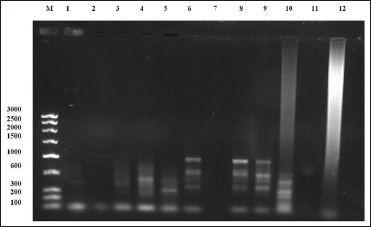 | Figure 9: Random amplified polymorphic DNA-polymerase chain reaction fragment patterns obtained by RAn-06 from tissues of Sperata seenghala preserved for 20 months. 1– 5 = tissues preserved in 8% formalin, 6– 9 = tissues preserved in 90% alcohol, 10– 12 tissues preserved in deep freezer. M is molecular marker (bp) of low range DNA ladder. [Click here to view] |
 | Figure 10: Random amplified polymorphic DNA-polymerase chain reaction fragment patterns obtained by RAn-05 from tissues of Sperata seenghala preserved for 28 months. 1– 4 = tissues preserved in 90% alcohol, 5– 8 = tissues preserved in 8% formalin, 10– 12 tissues preserved in deep freezer. M is molecular marker (bp) of low range DNA ladder. [Click here to view] |
4. ACKNOWLEDGMENTS
The authors wish to thank Director General, MPCST, Bhopal, for their help and encouragement during the work and financial support. The authors also wish to thank to laboratory mates for his help during this work.
5. REFERENCES
1. Chase MW, Hills HH. Silica gel: An ideal material for field preservation of leaf samples for DNA studies. Taxon 1991;40:215-20.
2. Klanten SO, Herwerden LV, Choat JH. Acquiring reef fish DNA sequences from formalin-fixed museum specimens. Bull Mar Sci 2003;73:771-6.
3. Martin C, Wasko AP, Oliveira C, Foresti F. Mitochondrial DNA variation in wild populations of Leporinus elongatus from the Parana River basin. Genet Mol Boil 2003;26:33-8.
4. Dawson MN, Raskoff KA, Jacobs DK. Field preservation of marine invertebrate tissue for DNA analyses. Mol Mar Biol Biotechnol 1998;7:145-52.
5. Dessauer HC, Cole CJ, Hafner MS. Collection and storage of tissues. In: Hillis DM, Moritz C, Mable BK, editors. Molecular Systematics. 2nd ed. Massachusetts: Sinauer Associates, Sunderland; 1996. p. 29-47.
6. Winsor L. Collection, handling, fixation, histological and storage procedures for taxonomic studies of terrestrial flatworms (Tricladida, Terricola). Pedobiologia 1998;42:405-11.
7. Fang SG, Wan QH, Fujihara N. Formalin removal from archival tissues by Critical Point Drying. Biotechniques 2002;33:604-11.
8. Koshiba M, Ogawa K, Hamazaki S, Sugiyama T, Ogawa O, Kitajima T, et al. The effect of formalin fixation on DNA and the extraction of high-molecular-weight DNA from fixed and embedded tissues. Pathol Res Pract 1993;189:66-72.
9. Green JE, Speller CF. Novel substrates as sources of ancient DNA: Prospects and hurdles. Genes 2017;8:1-26.
10. Janarthan S, Vincent S. Practical Biotechnology, Methods and Protocols. Vol. 13. India: University Press (India); 2007.
11. Laith AJ. The effect of formalin, alcohol and freezing on some body proportions of Alepes djeddaba Pisces: Carangid. Rev Biol Mar Ocean 2000;38:77-80.
12. Shields AP, Carlson SR. Effects of Formalin and alcohol preservation on length and weights of juvenile Sockeye Salmon. Alaska Fish Res Bull 1996;3:81-93.
13. Straube D, Juen A. Storage and shipping of tissue samples for DNA analyses: A case study on earthworms. Eur J Soil Biol 2013;57:13-8.
14. Rahman S, Daniel S, Hughes J. Development of an efficient method for producing high quality genomic DNA in Crustaceans. Asian J Anim Sci 2017;11:214-20.