1. INTRODUCTION
Banana is a major staple food, which provides not only calories but also other important components for human nutrition, such as vitamins, minerals, antioxidants, and prebiotics [1]. It is a fruit crop of global importance with an annual production of 102 million tonnes worldwide (http://faostat.fao.org). Banana follows ethylene-dependent ripening; hence, it falls in the category of climacteric fruit. In banana, different processes such as increase in the climacteric respiration, de-greening of peel, and softening of pulp are mainly coordinated by ethylene signaling. However, excessive softening of the fruit leads to the susceptibility to pathogens, which decreases the shelf-life of the fruit and results in huge post-harvest losses. Fruit softening is a vital indicator of the banana ripening process, which involves the disassembling of polysaccharides and is mainly regulated by several cell wall modification proteins such as pectate lyase (PL), polygalacturonase (PG), pectin methylesterases (PME) and xyloglucan endotransglucosylase/hydrolase (XTHs) [2,3]. Ethylene plays a key role in triggering the fruit ripening in climacteric fruit like bananas [4]. Interestingly, other phytohormones such as auxin and abscisic acid (ABA) are suggested to play a key role in promoting the fruit ripening process by cross talk with ethylene [5,6]. Previous studies by Inaba et al. [7] have clearly demonstrated that ethylene signaling was negatively controlled in banana pulp tissue, whereas positively controlled in banana peel [7]. Despite all these, the exact molecular mechanisms that coordinate the ethylene and other plant hormones, such as ABA and auxin signaling during the climacteric fruit ripening remain unclear.
The proteomic approach has become an important omics tool in fruit biology and its post-harvest research. In addition, proteomic studies can contribute to the understanding of fruit development and ripening through the identification of many metabolic and functional proteins that are operational in different tissue types during fruit development and ripening [1]. Therefore, a better understanding of the banana fruit ripening mechanism may help to develop strategies for reducing the post-harvest associated losses of the fruit crops in general and bananas in particular. Recently, proteome studies have been widely applied in investigating the molecular mechanism underlying the fruit ripening process in different fruits such as peach [8], pepper [9], apple [10], oil palm [11], and strawberry [12]. Previously, Hu et al. [13] have performed a spatiotemporal protein expression in peach endocarp and mesocarp during an early stage of development. Where they found that the protein abundance related to lignin and flavonoid pathways was found to be different for endocarp and mesocarp tissues during different developmental stages.
In an effort to decipher the regulatory mechanism of homogalacturonan (HG) genes during banana peel ripening Ning et al. [14] have performed a spatiotemporal gene expression study, suggesting that HG expression was found to be up-regulated during the ripening stage. Hitherto, very few spatiotemporal studies on gene/protein expression have been conducted on diverse fruits, to gain deeper molecular insight into the ripening process. However, those studies are confined to performing the expression analysis of a limited number of genes/ proteins that are related to specific metabolic pathways [14], whereas the role of other stage-specific or tissue-specific proteins in fruit development and ripening remains unexplored for many fruits, including bananas.
To date, most of the fruit proteome studies were restricted to a single tissue type and a limited number of development and ripening stages, whereas protein and their abundance in connection with different metabolic pathways related to fruit development and ripening are still far from fruition. In this regard, the present study provides an important insight into the molecular network during banana ripening and identifies proteins involved in the regulation of different metabolic pathways that could be helpful for reducing post-harvest loss in bananas. The present study provides an important insight into the molecular networks underlying banana fruit development and ripening. The objective of the study is (i) to decipher the proteome dynamic changes in a spatiotemporal manner in peel and pulp tissue of bananas using a high-throughput proteomics platform, (ii) to gain molecular insights of the proteins in relation to different metabolic pathways and their interactions in banana peel and pulp tissues from different developmental and ripening stages. Our study highlights the extensive peptide coverage and also resulted in the identification of tissue-specific proteins that are involved in different biological processes and metabolic pathways during banana development and ripening. Identified proteins could be highly useful in conducting further investigations to assign individual functional roles in fruit ripening and shelf life in bananas, thereby reducing post-harvest losses.
2. MATERIAL AND METHODS
2.1. Fruit Material
The cavendish banana fruit (Musa acuminata cv. Grand Naine) during different developmental stages, i.e., 40, 60, and 90-DAF were collected from OUAT, Bhubaneswar, Odisha. Banana fruits at the fully developed stage, i.e., 90-DAF were kept at ambient room temperature for further ripening for 6 and 12-DAR (days after ripening). Based on the uniformity in size, shape, color, firmness, visual defects, and appearance, fruits were selected for the experiment. Subsequently, the banana peel and pulp were separated at different developmental and ripening stages. The tissues were fine powdered with the help of liquid N2 and stored at −80°C until further use for protein extraction. Banana fruit tissues (peel and pulp) at different developmental and ripening stages were considered for the biometric, biochemical, and proteome analysis.
2.2. Biometric Assays
2.2.1. Measurement of fruit firmness
The fruit firmness was analyzed by puncturing the fruit using a penetrometer (model no. FR-5120, Lutron, USA) with a 6 mm plunger tip. A small slice of banana fruit skin was torn off, and firmness was determined from three replicated fruits with five different points per fruit. The maximum amount of force required to penetrate the plunger tip into the banana fruit was recorded and expressed in Newton (N).
2.3. Biochemical Assays
2.3.1. Estimation of starch
Banana fruit tissues at different developmental (40, 60, and 90-DAF) and ripening stages (6 and 12-DAR) were considered for the starch estimation. Starch estimation was performed by following the procedure of Shafiee et al. [15]. The experiment was performed by taking three biological replicates for each of the stages.
2.3.2. Estimation of total sugar content
Total sugar estimation was performed by following the method of Franscistt et al. [16] and triplicated readings were recorded from three independent biological replicates for each stage.
2.4. Protein Extraction
Molecular-grade chemicals were used for the protein isolation. Proteins were extracted from the banana tissues (peel and pulp) at different developmental and ripening stages by following the phenol extraction method described by Carpentier et al. [17] with slight modification.
2.4.1. Reconstitution of protein pellet in 1×PBS buffer
30 mg of dry protein pellet samples were dissolved in 300 µl of 1 × PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4) and the mixtures were vortexed briefly for 5 minutes and centrifuged the samples at 12,000 × g for 5 minutes at 4°C. The clear supernatant was separated, transferred to a new 1.5 ml Eppendorf tube, and stored at −80°C for further use.
2.5. Quantification of Protein Samples
Protein concentration was estimated using Quick Start 1× Bradford assay (Bio-Rad, USA, 5000205) reagent, using BSA as standard. Protein samples were considered duplicates for each of the stages for quantification. The absorbance value of unknown protein samples was measured at 595 nm. Based on the absorbance and concentration of BSA, a standard curve was plotted.
2.6. SDS-PAGE Separation of Proteins
During SDS-PAGE analysis, protein samples (100 μg) were dissolved in loading buffer (0.5 M tris-HCl [pH 6.8], 10% SDS, glycerol, and β-mercaptoethanol) and the mixture was incubated at 95°C in the water bath for 4 minutes. The protein ladder (Bio-Rad, USA,1610317) with a molecular mass molecular weight (Mw) range from 6.5 to 200 kDa was loaded in the first well followed by protein samples and resolved by 12% polyacrylamide gel, which was then stained with GelCode blue stain reagent (Pierce, USA, 24590).
2.7. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) Analysis for Protein Identification and Quantitation
Proteins were reduced, alkylated, and digested at 37°C in ammonium bicarbonate and with trypsin as described by Ray et al. [18]. Peptides were desalted using spin columns and subjected to Orbitrap fusion mass spectrometry. Furthermore, protein identification and quantification were done as per the method described in the above-cited reference.
2.8. Data Analysis
Proteome discoverer version 2.2 (Thermo Fisher Scientific) was used for processing the raw data files generated from the mass spectrometry. Spectra (MS2) were searched using Mascot search engine (version 2.6.0) against the M. acuminata database fasta from the UniProt sequence tools. The parameters for protein database search were followed as per the instructions of bioinformatics tools. The proteins with a false discovery rate of 1% were considered for protein identification.
2.9. Protein–Protein Interactions
Protein–protein interactions (PPIs) network was analyzed by using GeneMania.org by following the method of Warde-Farley et al. [19]. While the genes from M. acuminata were mapped to GeneMania, these interologs were then checked directly using String-db.org (www.string.embl.de) in the Musa database, which unfortunately maps to very few. The pulp 6 and 12 DAR differentially expressed proteins were mapped to GeneMania Arabidopsis thaliana to derive the orthologous interacting pairs (interologs). While betweenness, closeness, and centralities could form the other ranking coefficients, the clustering coefficient would yield more distinct interaction partners.
2.10. Functional Categorization
Proteins identified through mass spectrometry were functionally categorized based on the literature on different fruits connected with the ripening process. The common and unique proteins were analyzed using Venny: https://bioinfogp.cnb.csic.es/tools/venny/ and further pathway maps were checked between them.
3. RESULTS
Morphological changes were observed in banana fruit during different developmental (40, 60, and 90-DAF) and ripening stages (6 and 12-DAR). The visual appearance of fruits clearly differentiates the development and ripening stages of bananas. In the case of 90-DAF (pre-climacteric stage), the fruits are bigger in size with pedicel portion, and slight ridges and color of the fruit appear to be green with slight traces of yellow, compared to other stages. Whereas, in the case of 12-DAR (climacteric stage), no ridges could be visualized and the pedicel portion turned black and dry, the color of the fruit was brown with patches, and the texture of the fruit was soft compared to 90-DAF fruit (Fig. 1). The change in the color of the fruit during the progression of ripening is due to the pigmentation and degradation of chlorophyll. Fruit firmness slightly increased during developmental stages, reached a maximum at 90 DAR, and decreased at 6 and 12 DAR (Fig. 2).
 | Figure 1. Visual appearance of banana cv. Grand Naine at different developmental and ripening stages. Fruit sampling was done at 40-DAF (A), 60-DAF (B), 90-DAF (C), 6-DAR (D), 12-DAR (E) and considered for the biometric, biochemical and proteome analysis. [Click here to view] |
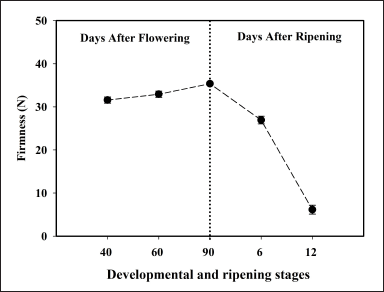 | Figure 2. Pulp firmness at different developmental and ripening stages. Each value represents the mean of three biological replications of three fruits analyzed at each ripening stage and vertical error bars represent the standard deviation (±SD). [Click here to view] |
In the present study, banana fruit tissue displayed a high starch content at developmental stages, i.e., 40, 60, and 90-DAF, and was found to be decreased during different stages of ripening, i.e., 6 and 12-DAR (Fig. 3A), this explains the starch hydrolysis and synthesis of sugars during ripening. In addition, patterns of the starch content are consistent with the expression of starch biosynthesis pathway-related proteins that were identified through mass spectrometry (Fig. 6C). Interestingly, in the present study, the total sugar content showed an increasing pattern from banana developmental to ripening stages, indicating that sugar accumulation is high during the onset of ripening (Fig. 3B). Furthermore, the pattern of total sugar content was similar to the expression profiles of sugar metabolism proteins (Fig. 6D), suggesting that starch breakdown into to simple sugars resulted in textural changes during banana fruit ripening.
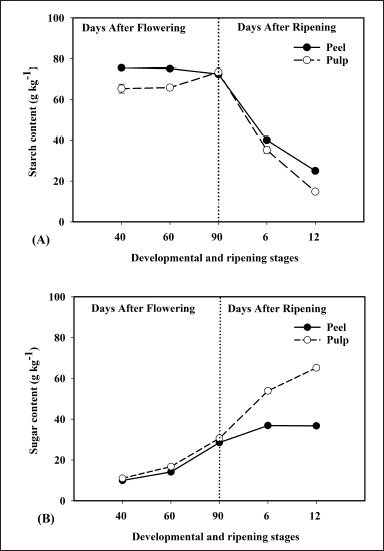 | Figure 3. Represents the biochemical changes in peel and pulp tissue of banana fruit during different developmental and ripening stages. Starch content (A) and total sugar (B) of banana peel and pulp tissues at different developmental and ripening stages, and expressed in g kg-1. Each value represents the mean of three biological replications of three fruits analyzed at each ripening stage and vertical error bars represent the standard deviation (±SD). [Click here to view] |
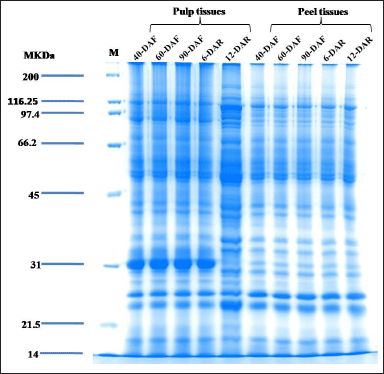 | Figure 4. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis profile showing separation of proteins from banana pulp and peel tissues of different developmental and ripening stages by using phenol-based extraction method. A known amount of protein (100 µg) was loaded in each lane and resolved on 12% SDS-PAGE followed by Colloidal Coomassie blue staining. M: SDS-PAGE Mw standards (KDa), DAF: Days after flowering, DAR: Days After Ripening. [Click here to view] |
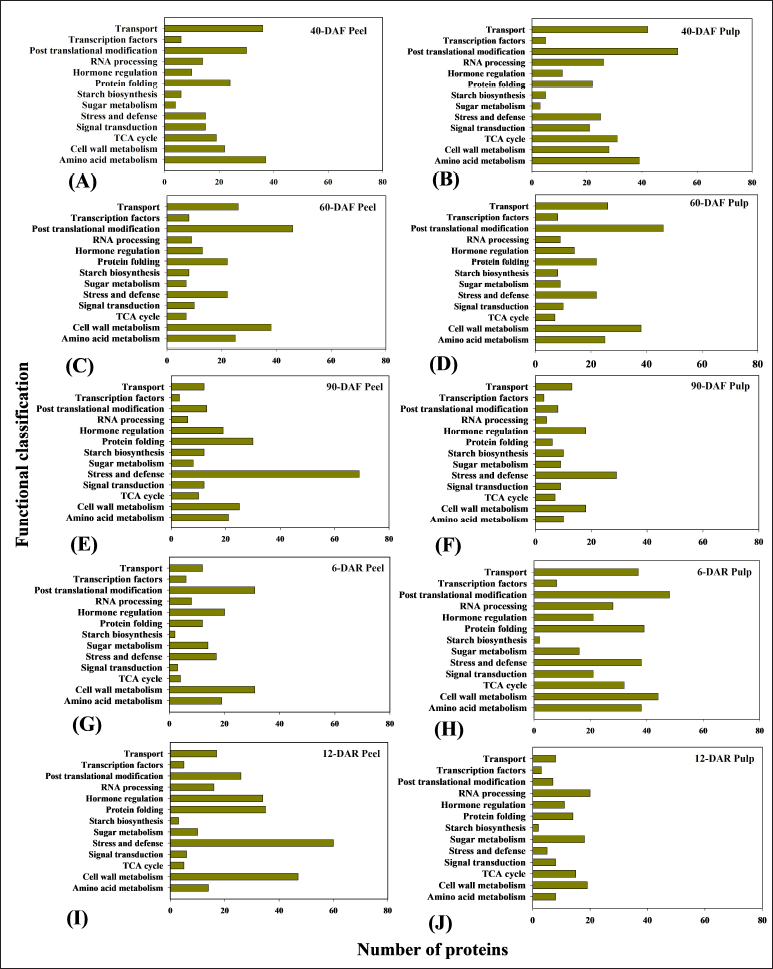 | Figure 5. Functional categories of the identified proteins from banana fruit tissues at different developmental and ripening stages represented as 40-DAF peel (A), 40-DAF pulp (B), 60-DAF peel (C), 60-DAF pulp (D), 90-DAF peel (E), 90-DAF pulp (F), 6-DAR peel (G), 6-DAR pulp (H), 12-DAR peel (I), 12-DAR pulp (J), through Orbitrap fusion mass spectrometry (mass spectrometer combines best of quadrupole, orbitrap and linear ion trap; tribrid) analysis. [Click here to view] |
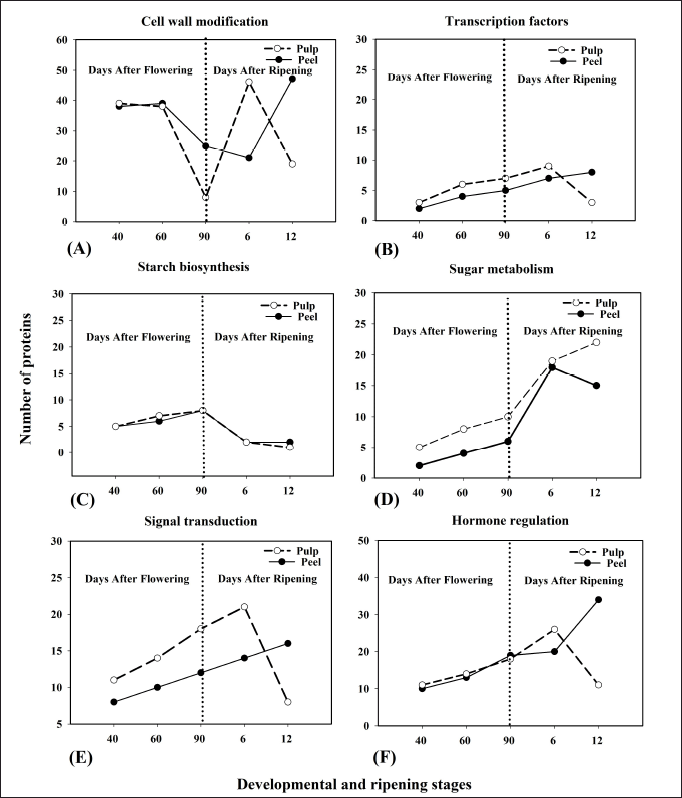 | Figure 6. Spatiotemporal expression of functionally categorized proteins those are identified from banana peel and pulp tissues at different developmental and ripening stages. Cell wall modification (A), TFs (B), starch biosynthesis (C), Sugar metabolism (D), signal transduction (E), hormone regulation (F). [Click here to view] |
The proteins from banana peel and pulp at different developmental and ripening stages were extracted by the phenol extraction method (Fig. 4). The SDS-PAGE profiles of banana peel and pulp at different developmental and ripening stages exhibited the qualitative difference in banding patterns. The majority of the proteins were separated at a range of 14 to 150 kDa. Strong and intense protein bands were observed in pulp tissue at 31 kDa in developmental (40, 60, and 90-DAF) and ripening stage (6-DAR) due to the presence of abundant protein (except for 12-DAR pulp), while in the case of peel proteins at 31 kDa showed light bands. We assume that 31 kDa protein may play a role in starch breakdown and sugar synthesis in pulp tissue. Interestingly, the protein profiles of 12-DAR pulp exhibited abundant protein banding patterns between the molecular weight (Mw) range of 70 to 116.25 kDa, we anticipate that these proteins may have a role in pulp softening banana pulp softening. We have also identified low Mw proteins with light bands ranging from 14 to 21.5 kDa in peel and pulp tissues of different developmental and ripening stages. Overall, the proteins were well separated without the presence of any smears or degradation.
Orbitrap fusion mass spectrometry coupled with nano LC-MS/MS allowed the identification of 5,701 and 6,959 proteins in banana peel and pulp tissues, respectively, from different developmental and ripening stages. The functional role was assigned to a total of 1,646 and 1,806 proteins in banana peel and pulp tissues based on the available literature and protein database search (Suppl Tables 1–10). Furthermore, a total of 1,176 proteins from peel and 1,209 proteins from pulp tissues were selected and presented in Figure 5. Identified proteins were tabulated with respective accession number, sum peptide score, peptide coverage (%), amino acid, Mw in kDa, unique peptides, and peptide sequence (Tables 1–10) (Supplementary file).
The majority of the identified proteins were involved in diverse metabolic pathways during banana fruit ripening, i.e., starch and sugar metabolism, cell wall metabolism, hormone regulation, post-translational modifications, signal transduction, and transcription factors (TFs) (Fig. 5A–J). Proteins identified through mass spectrometry-based platforms exhibited varying abundance between peel and pulp tissues at different developmental and ripening stages. In contrast, proteins related to cell wall modification and hormone regulation exhibited high abundance in the case of 12-DAR peel when compared to other developmental and ripening stages. Sugar metabolism-related proteins exhibited high abundance in the 12-DAR pulp. The overall functional classification is shown in Figure 5A–J.
Figure 6 represents the expression pattern of functionally categorized proteins based on their number that are identified in banana peel and pulp tissues at different developmental and ripening stages. Identified proteins are involved in different metabolic pathways such as cell wall modification, TFs, starch biosynthesis, sugar metabolism, signal transduction, and hormone regulation (Fig. 6A–F). The starch biosynthesis protein pattern of peel and pulp exhibited high abundance during different developmental stages, i.e., 40-DAF, 60-DAF, and 90-DAF, and gradually the trend decreased at the ripening stage, i.e., 6-DAR and 12-DAR (Fig. 6C). Sugar metabolism related category of proteins number gradually increased during progression of ripening and showed maximum increase at 12-DAR pulp. However, in the case of 12-DAR peel, slight decreases in the number of sugar metabolism proteins were observed (Fig. 6D). The trend of the TF and signal transduction proteins was similar in peel and pulp tissues at different developmental and ripening stages of the banana (Fig. 6B and E). Initially, the number of TF proteins in the peel gradually increased at 40-, 60-, 90-DAF, and 6- and 12-DAR. However, TF proteins in pulp showed an increase in the pattern at 40-, 60-, 90-DAF, and reached a maximum at 6-DAR but decreased at 12-DAR. The number of cell wall modification proteins in peel tissue gradually decreased from 40-DAF to 6-DAR but sharply increased at 12-DAR. However, the same category of proteins in pulp showed a decrease in the pattern from 40- to 90-DAF, and later showed a sharp increase at 6-DAR but decreased at 12-DAR (Fig. 6A). Among all the developmental and ripening stages of banana fruit, 12-DAR peel exhibited the highest number of proteins associated with hormone regulation (Fig. 6F).
We observed varied spatiotemporal expression of the proteins that were regulating different metabolic pathways in banana peel and pulp tissues during developmental and ripening stages (Fig. 7A–J). An increase in the expression pattern of α-mannosidase (α-man) was observed in the pulp tissue from 40-DAF to 6-DAR (4-fold), but sharply decreased at 12-DAR pulp. Whereas, in the case of peel tissue, the expression pattern of α-Man protein gradually increased from 40- to 90-DAF, but decreased in the later stages of ripening, i.e., 6- and 12-DAR (Fig. 7A). β-galactosidase (β-gal) protein showed an increase in the abundance in both peel and pulp at 40-, 60- and 90-DAF, but decreased at 6-DAR peel and pulp. Whereas, the same protein was expressed at 12-DAR peel but decreased expression at 12-DAR pulp (Fig. 7B). The abundance of the PE proteins in the peel showed declined at 60-DAF and sharply increased at 90-DAF, 6, and 12-DAR. In the case of pulp, the expression of PE proteins was found to decrease from 40-DAF to 6-DAR, but a sharp increase in the pattern was observed at 12-DAR (Fig. 7C). Our result showed that the XTH protein, which is involved in cell wall modification, was more abundant in peel tissue at 12-DAR (3-fold) than at other developmental stages, i.e., 40, 60, and 90-DAF and 6-DAR. In the case of pulp, the pattern of XTH gradually increased at developmental stages (40 to 90-DAF) but slightly decreased at 6 and 12-DAR (Fig. 7D).
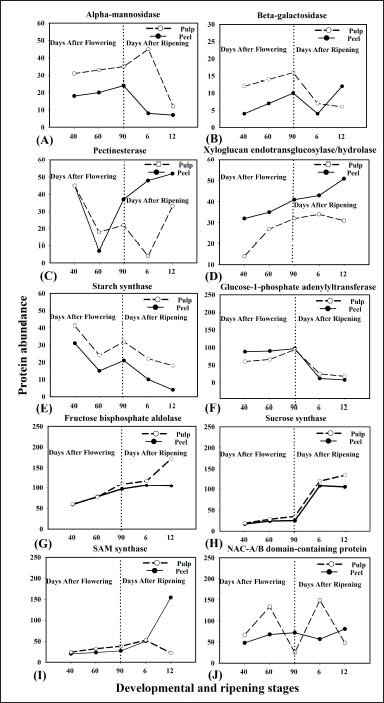 | Figure 7. Spatiotemporal expression analyses of individual proteins from banana peel and pulp tissues at different developmental and ripening stages. Proteins involved in different metabolic and regulatory pathways during developmental and ripening stages of bananas were found to be differentially expressed. α-man (A), β-gal (B), Pectinesterases (PE) (C), XTH (D), SS (E), G-1P adenyltransferase (F), Fructose bis-phosphate aldolase (G), Sucrose synthase (H), SAM synthase (I), NAC-A/B domain-containing protein (J). Cell wall modified proteins: Pectinesterases, α-man, XTHs and β-gal. Starch metabolism proteins: SS and G-1P adenyltransferase. Sugar metabolism proteins: Sucrose synthase, fructose bis-phosphate aldolase. TFs: NAC-A/B domain containing proteins. Ethylene biosynthesis: SAM synthase. [Click here to view] |
Starch biosynthesis-related proteins such as starch synthase (SS) showed an increase (2-fold) in the abundance in both peel and pulp at the initial stage of fruit development, i.e., 40-DAF, but decreased with the progression of banana ripening (Fig. 7E). Glucose-1-phosphate (G-1P) adenylyltransferase protein which is also related to starch biosynthesis were found to be increased in both peel and pulp during developmental stages of banana, i.e., 40,60 and 90-DAF, but decreased during ripening stages, i.e., 6 and 12-DAR (Fig. 7F).
Sugar metabolism-related proteins such as sucrose synthase and fructose-bis-phosphate aldolase showed an increase in the abundance of pulp during the ripening stage, i.e., 12-DAR. The expression pattern of sucrose synthase and fructose-bisphosphate aldolase proteins in pulp gradually increased from 40-DAF to 12-DAR, but slightly decreased at 12-DAR peel (Fig. 7G–H).
S-adenosylmethionine synthase (SAM-synthase) proteins which are known to be involved in ethylene biosynthesis gradually increased in both peel and pulp from 40-DAF to 6-DAR, but later showed a decline in the expression in 12-DAR pulp. The highest expression of SAM synthase protein was observed in 12-DAR peel (3-fold) (Fig. 7I).
NAM, ATAF1/2 and CUC2 (NAC) A/B domain-containing protein which is involved in the regulation of ethylene signalling exhibited 1.5-fold increase in the expression pattern in peel tissue throughout the developmental and ripening stages, except at 6-DAR where it was slightly declined. In the case of pulp, a sharp increase in the expression (2-fold) was observed during the early stage of fruit development (60-DAF) and ripening (6-DAR). However, a sharp decline in the expression pattern of protein was observed at fully developed (90-DAF) and later stages of ripening (12-DAR) (Fig. 7J).
Identified proteins (presented in bold letters), known to regulate different metabolic pathways during the banana fruit development and ripening process, are represented schematically (Fig. 8). Interestingly, TFs-related proteins, i.e., bZIP, NAC-A/B, and APETALA2/ethylene response factor (AP2/ERF) are known to regulate different metabolic pathways such as hormone regulation and sugar metabolism which resulted in cell wall loosening during ripening. Ethylene is suggested to cross-talk with other phytohormones such as auxin and IBA for triggering the fruit ripening process (Fig. 8). In contrast, some of the proteins were found to be present at specific stages and tissue types of banana, which were represented in the numerical format.
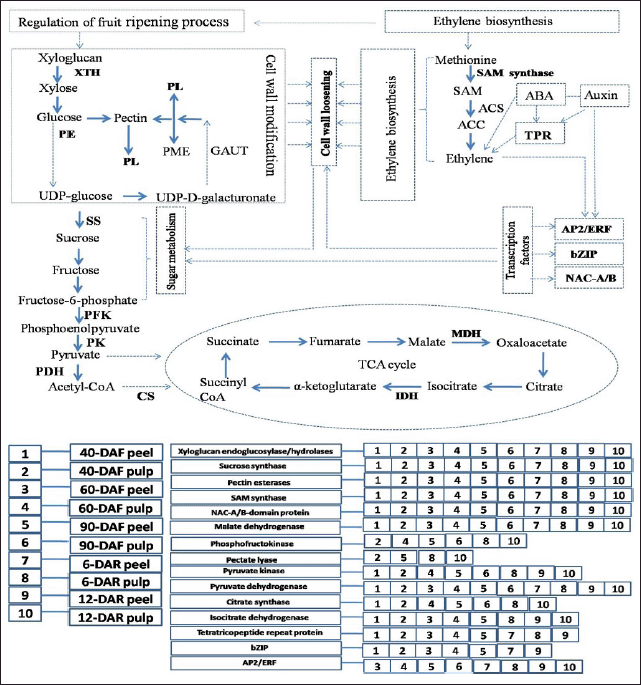 | Figure 8. Schematic overview of the proteins and their regulation in different pathways during banana fruit development and ripening process. Identified proteins were represented in bold letters which were involved in cell wall modification, sugar metabolism, TCA cycle, TFs, ethylene biosynthesis and hormone regulation. Cell wall modification proteins: XTH, PL, pectin esterase (PE), sugar metabolism proteins: sucrose synthase, TCA cycle: phosphofructokinase (PFK), pyruvate kinase (PK), pyruvate dehydrogenase (PDH), citrate synthase (CS), isocitrate dehydrogenase (IDH) and MDH, Ethylene biosynthesis: SAM synthase, TFs: AP2/ERF, bZIP and NAC-A/B, Auxin and ABA signalling: TPR protein. Identified proteins were present in specific stages and tissue types were assigned different digits/codes for banana pulp and peel tissues of different developmental and ripening stages (1–10). [Click here to view] |
Venn diagrams representing the common and distinct proteins through various comparisons: (A) 40-DAF pulp, 60-DAF peel, 40-DAF peel, and 60-DAF pulp; (B) 40-DAF pulp, 90-DAF pulp, 40-DAF peel, and 60-DAF pulp; (C) 6-DAR pulp, 12-DAR peel, 6-DAR peel, and 12-DAR pulp, are depicted in Fig. 9A and C. In the case of 40-DAF pulp and 60-DAF peel, a total of 6 (42.9%) and 2 (14.3%) unique proteins, with no common proteins. A total of 2 (14.3%) and 1 (7.1%) distinct proteins, without having overlapping proteins were identified in 40-DAF peel and 60-DAF pulp, respectively. Further, a total of 2 (11.1%) and 1 (5.6%) proteins were unique for 40-DAF pulp and 90-DAF pulp. Six (33.3%) and 1 (5.6%) distinct proteins, without sharing any overlapping proteins were identified in 40-DAF peel and 60-DAF pulp, respectively. In the case of 6-DAR pulp and 12-DAR peel, a total of 16 (44.4%) and 7 (19.4%) distinct proteins were present without sharing any common proteins. Likewise, unique proteins of 3 (8.3%) and 2 (5.6%) were present in 6-DAR peel and 12-DAR pulp, suggesting the presence of distinct unique proteins may have a regulatory role at specific stages and tissue types.
 | Figure 9. Venn diagram exhibiting the commonality and uniqueness of identified proteins in different tissues and stages of banana fruit. The comparisons were made between 40 and 60-DAF peel and pulp tissue (A), 40 and 90-DAF peel and pulp tissue (B), 6 and 12-DAR peel and pulp tissue (C). Top 10 highly expressed proteins were considered for the comparison. [Click here to view] |
Figure 10 presents the PPI network analysis of peel and pulp tissues of different developmental and ripening stages by using gene Mania. Two glycolysis-related proteins, i.e., enolase and phosphoglucomutase (PGM), were found to interact more with other proteins. PGM acts as an important regulatory enzyme in cellular glucose utilization and energy homeostasis process during ripening and also it participates in sugar metabolism. Whereas, enolase is involved in the energy metabolism and carbon flow process during fruit ripening.
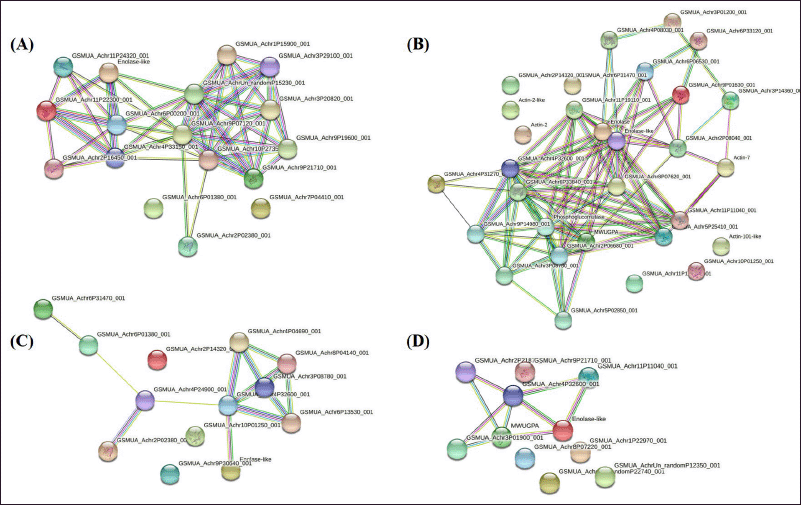 | Figure 10. The PPI network is simulated by STRING. PPI is presented for the identified proteins (top 20) in bananas during different developmental and ripening stages. Arabidopsis thaliana network database and a confidence level of 0.4 were used for the analysis parameters. Different line colours represent the types of evidence used in predicting the associations. The comparisons were made between 6 and 12 DAR peel (A), 6 and 12 DAR pulp (B), 40, 60 and 90-DAF peel (C) and 40, 60 and 90-DAF pulp (D). [Click here to view] |
4. DISCUSSION
Ethylene cross talk with other hormones such as ABA for the promotion of the fruit ripening process by stimulating the signaling of several ethylene components, resulting in ethylene-induced fruit softening [5]. However, the molecular mechanisms that coordinate auxin, ABA, and ethylene signaling during the fruit ripening process remain unclear. Fruit softening depends on the alteration of the cell wall structural properties, with massive depolymerization/solubilization of cell wall components such as polysaccharides (pectins, cellulose, and hemicelluloses), lignin, and proteins [1,20].
In the present study, the decline in the starch content with the rise in sugar accumulation, clearly demonstrates the starch hydrolysis and synthesis of sugars during banana ripening. This study focused on the spatiotemporal protein expression of banana fruit from early development to late ripening stage. In addition, differentially expressed proteins that regulate different metabolic pathways during banana fruit development and ripening processes were described.
4.1. Proteins Related to Starch and Sugar Metabolism
During the ripening, the fruit undergoes several biochemical and physiological changes that result in starch breakdown to sugar or sucrose accumulation [21]. Starch is a polymer of glucose-1-phosphate (G-1P), linked by an α-1,4-glycosidic bond, which is usually synthesized for energy storage during fruit maturation. Through the proteome analysis, Chin et al. [22] have identified α-1,4-glucan phosphorylase proteins, which exhibited high abundance during the unripe stage of mango pulp. Interestingly, in our study, α-1,4-glucan phosphorylase proteins (#2) were increased by 1.5-fold in the pulp tissue of fully developed fruit and exhibited less abundance in the rest of the developmental and ripening stages, suggesting that the α-1,4-glucan phosphorylase activity reaches a maximum in fully developed fruit thereby accumulation of high starch content in the pulp tissue. SS protein which participates in the starch biosynthesis process was decreased in banana pulp during ripening [1]. The spatiotemporal expression analysis revealed an increase in the protein expression pattern by 2-fold in both peel and pulp during the early stages of fruit development. However, the expression pattern of the same protein displayed a decreased pattern in both the tissues during the later stage of development and ripening, and our results are in consistent with the previous reports [1].
SuSy is actively participates in the carbohydrate polymers synthesis, i.e., starch or cellulose, or in the generation of active compounds which helps in fruit development [23]. In this regard, Tian et al. [23] have identified SuSy proteins in kiwi fruit tissues (exocarp) at different developmental and ripening stages. Furthermore, during the onset of kiwi fruit ripening, SuSy was found to be up-regulated [23]. In the present study, SuSy exhibited increased protein abundance in both peel and pulp tissues across the developmental and ripening stages of bananas. However, a sharp rise (2-fold) in SuSy was observed during the ripening stages of banana pulp tissue, suggesting that sugar synthesis is highly active in pulp tissue during fruit ripening.
Fructose-bisphosphate aldolase has a key role in sugar metabolism and it is involved in the regulation of the sink metabolism of the fruit tissue during the ripening process [24,25]. Previous reports have observed a significant increase in the fructose-bisphosphate aldolase proteins in melon and blueberry ripening [24,25], suggesting that fructose-bisphosphate aldolase may have a key role in sucrose metabolism at ripening [24,25]. In this study, fructose-bisphosphate aldolase expression gradually increased (2-fold) in peel tissue from developmental to early ripening stages. Furthermore, the protein expression of fructose-bisphosphate aldolase gradually increased (2-fold) in the pulp tissue during fruit development and ripening, suggesting its active role in the synthesis and accumulation of sugars during fruit ripening.
4.2. Proteins Related to Hormone Regulation
Plant hormones are widely known to be major regulators of fruit development and ripening. In this regard, ethylene has a major role in triggering the expression of cell wall modifying enzymes such as PME, PL, and PG, that catalyze the cell wall pectin depolymerization, resulting in the gradual fruit softening [4,26,27]. SAM-synthase proteins are known to be involved in ethylene and polyamine biosynthesis and they catalyze SAM from methionine during fruit ripening [28]. A major portion (80%) of the cellular methionine is converted to SAM by the action of SAM-synthase and ATP [29].
Previous studies have reported that the abundance of SAM synthase proteins was increased in cherry tomato pericarp and peach mesocarp during ripening [13,29]. It was also observed that the abundance of SAM-synthase was relatively higher in the early stage of tomato ripening compared to the later stage ripening [29]. Interestingly, in the present, study the expression pattern of SAM-synthase protein gradually increased in both peel and pulp from the developmental to the early ripening stage, but at later stages of ripening its expression declined in the pulp. In averse, a sharp rise (3-fold) in the expression of SAM synthase protein was observed in the peel compared to pulp tissue, reflecting the abundant expression of SAM synthase in the peel tissue. We speculate that the site of synthesis of ethylene is in the peel tissue and later it translocates to the pulp tissue to trigger the ripening process. In this context, our results are in agreement with the previous studies on different fruits that the SAM-synthase may play a key role in triggering the fruit ripening process by converting S-adenosyl-L-methionine to 1-aminocyclopropane-1-carboxylic acid (ACC) during ethylene biosynthesis process.
ARF-GAP mediates plant growth and root development by regulating auxin levels which were previously reported in rice and Arabidopsis [30,31]. The auxin and ethylene pathways cooperatively regulate a variety of developmental processes in plants. The auxin is capable of inducing ethylene biosynthesis and signaling and these two hormones interact reciprocally [32]. However, the role of ARF-GAP proteins in fruit ripening is still not clear. The present study emphasizes that the expression level of ARF-GAP protein was abundant in pulp (4-fold) tissue compared to peel tissue during the ripening stage of banana. Furthermore, our study speculates that the ARF-GAP proteins may have a functional role in mediating the auxin efflux cross talk with ethylene signaling for triggering the fruit ripening in bananas.
Tetratrico-peptide repeat (TPR) domain proteins are involved in auxin and ABA signaling. In Arabidopsis, it was reported that the ETO1 (ETHYLENE-OVERPRODUCER1) protein negatively regulates ethylene biosynthesis in seedlings through direct interaction of its TPR domains with an ACC synthase isoform [33]. However, the specific role of TPR proteins in fruit ripening is yet to be explored. It was observed that ABA regulates the ethylene biosynthesis and ABA-ethylene interaction leads to the triggering of the fruit ripening process [6]. Despite all these, the exact molecular mechanism exploring the interaction between ABA and ethylene during fruit ripening has not yet been reported. In the present study, TPR_REGION domain protein which is involved in auxin and ABA signaling gradually decreased in the pulp tissue at different stages of fruit development and in the early stage of ripening, but the same protein was found to be absent in pulp tissue of fully developed fruit and at the later stage of ripening. On the other hand, the expression pattern of TPR_ REGION domain protein in the peel tissue exhibited a sharp increase (2-fold) at later stages of fruit ripening. The modulation of SAM-synthase, ARF-GAP, and TPR proteins in different tissues of banana indicates the coordinated action of ethylene, auxin, and ABA hormones and their positive and negative regulation in promoting fruit growth, development, and ripening.
4.3. Proteins Related to Cell Wall Modification
Cell wall disassembly promotes banana ripening by triggering the breakdown of multiple polysaccharides networks, with the help of cell wall degrading proteins such as PL, PG, PME, and others [3]. The PL proteins involved in the maceration and soft-rotting of fruit tissue and up-regulation of PL in peel and pulp tissues were observed in different studies during fruit ripening [1,20,34,35]. In the present study, a high abundance of PL proteins in the peel and pulp was observed at 12-DAR, suggesting that it could play a key role in the degradation of pectin during banana ripening.
XTH is another class of cell wall modifying proteins that contribute to cell wall integrity maintenance by endotransglucosylase, and weakening by hydrolase activity during fruit ripening. During fruit ripening, xyloglucan hydrolase mediates the cell wall expansion process [1,34]. Importantly, XTH proteins were involved in the regulation of auxin and ethylene-mediated signaling and post-harvest fruit softening [34]. Ten XTH proteins, which were consistent with the degradation of cell wall components, were identified in the peel and pulp tissue of banana fruit at the ripening stage [36,34]. Further, Bhuiyan et al. [1] have identified eight up-regulated cell wall metabolism proteins such as XTH in the pulp tissues of banana fruit at the ripening stage. Another study by Kok et al. [11] has identified five numbers of XTH proteins in oil palm which were found to be up-regulated during ripening stages. In the present study, XTH protein expression in peel gradually increased during development and exhibited a sharp increase at ripening (3-fold), whereas in the pulp tissue XTH protein expression gradually increased from the developmental to ripening stage. Our result postulates that XTH may play an important role in banana peel softening during ripening by exhibiting hydrolyzing activity. α-Man is a fruit ripening-specific N-glycan processing enzyme, which is involved in the fruit softening process. The RNAi mediated suppression of α-Man genes, resulted in extended shelf-life for days 30 in tomato [37] and days 7 in capsicum [38]. Interestingly, in the present study, an increase in the expression pattern of α-Man was observed in the pulp tissue during development and early ripening (up to 4-fold), but sharply decreased at later stages of ripening in the pulp. In the case of peel tissue, the expression pattern of α-Man protein gradually increased during development but decreased in the later stages of ripening, disclosing its role in hydrolyzing the cell wall components, thereby increasing the fruit softening.
PE plays an important role in the cell wall metabolism during fruit ripening. This protein is known to extensively decrease the rigidity of the cell wall structure and solubilization of pectins during fruit softening [3]. PE proteins were identified in pulp tissues of jujube fruit at two mature stages and pulp tissues of papaya at ripe and unripe stages [39]. Furthermore, the expression of PE proteins was coherent with the degradation of cell wall components in peel and pulp tissue of banana at ripening [36,34]. Similarly, Xiao et al. [35] have identified two up-regulated PE proteins in banana peel tissues at the climacteric stage. In the present study, the abundance of PE proteins was noticed in both peel and pulp at the initial stage of fruit development. However, a sharp increase (1.5-fold) in the pattern of PE was observed in peel tissues at ripening stages. On the other hand, the expression pattern of PE proteins in the pulp decreased significantly but increased during the later stage of ripening. It suggests that the regulatory mechanism of PE is different for the peel and pulp tissues of bananas during fruit development and ripening. In addition, high expression of PE proteins in peel during the ripening stage could indicate that these proteins may play a key role in fruit softening by solubilizing the cell wall pectins.
β-gal is a cell wall modification protein involved in debranching pectin and supplementing the depolymerization process during fruit softening [22,40]. It is part of a wide family of glycosyl hydrolases known to remove β-galactosyl residues from the pectin side chain during fruit softening [40]. Chin et al. [22] have reported that β-gal increased in the expression during the ripening stage of mango pulp. Further, Niu et al. [41] have revealed that β-gal was up-regulated in cracking Akebia trifoliata fruits. In this study, β-gal protein expression gradually increased in both peel and pulp tissue during fruit development, but exhibited decreased abundance during the ripening. However, in the case of peel the expression pattern of β-gal showed an increase in the abundance at the later stage of ripening, which provides evidence that the β-gal actively participates in peel tissue during ripening by exhibiting the high depolymerization activity, thereby promoting the fruit softening.
4.4. Proteins Related to TFs
TFs play a key role in the regulation of fruit development and ripening [20]. In this regard, the AP2/ERF is a major TF involved in the modulation of the ethylene signaling pathway during the fruit development and ripening process [20,42]. iTRAQ and Q exactive mass spectrometry-based proteome analysis revealed that AP2/ERF proteins were found to be up-regulated in melon flesh during the developmental stage [43]. In the present study, AP2/ERF protein expression was noticed during later stages of fruit development; the expression was gradually increased and reached the maximum expression at later stages of ripening in the peel tissue. However, in the case of pulp, the expression pattern of AP2/ERF slightly declined during the later stage of ripening. The trend was similar to that of the expression of SAM-synthase suggesting that AP2/ERF proteins may have a role in the ethylene signaling pathway during banana fruit ripening.
The leucine-zipper (bZIP) protein plays a key role in various biological processes such as synthesis of volatile compounds, floral induction, and seed maturation. However, proteome studies to elucidate the regulatory mechanism of bZIP proteins in fruit development and ripening are yet to be explored fully. The bZIP proteins were found to be up-regulated significantly in the banana peel during ripening, and also suggested to be involved in the regulation of volatile aroma production during ripening [20]. In the present study, the expression pattern of bZIP protein in the peel tissue was induced at a later stage of ripening, but absent in the early stage of ripening. Two numbers of bZIP proteins were increased (2-fold) in the pulp tissue at the early-stage ripening, when compared to other developmental and ripening stages. In addition, our results are in consistent with the previous studies that bZIP proteins may play a crucial function in the synthesis of volatile compounds during ripening.
NAC proteins may have a major function in the regulation of phytohormone signaling (ethylene, ABA, and auxin) and also involve in pigment accumulation, floral development, and fruit softening [44,45]. Yun et al. [20] have identified NAC-domain proteins, which were significantly up-regulated in banana peel tissue during ripening. Interestingly, in the present study, a sharp rise in the expression pattern of NAC-A/B domain containing protein was observed in the peel tissue. In the case of pulp tissue, the expression of NAC-A/B-domain containing protein was increased during early developmental and early ripening stages. However, its expression is reduced in the pulp during fully developed and later stages of ripening. Our result speculates that the NAC-A/B domain-containing protein may participate in the banana fruit ripening process by interacting with the ethylene signaling pathway. The trend of the expression pattern of AP2/ERF, bZIP, and NAC-A/B domain-containing proteins was found to be similar to that of SAM synthase protein expression, suggesting that these proteins may have a key role in controlling the regulatory process of ethylene biosynthesis and signaling during ripening. In addition, these proteins may also regulate ethylene synthesis in a spatiotemporal and tissue-specific manner.
4.5. Proteins Related to Carbon and Energy Metabolism
Malate dehydrogenase (MDH) is a key protein responsible for malate oxidation process during fruit ripening and also participates in carbon fixation [46]. MDH is involved in the TCA cycle and is also suggested to play a key role in facilitating the exchange of metabolites between cytoplasm and organelles. Yu et al. [47] have revealed that MDH modulates the accumulation of carbohydrates by adjusting the carbohydrate allocation from sources to sink during apple fruit ripening. MDH protein has been known to regulate photosynthetic activity and anthocyanin biosynthesis in plum fruit [48]. Interestingly, in the present study, MDH protein in both peel and pulp tissue showed an increase in the expression during the initial stage of fruit development, but exhibited decreased expression during ripening. Meanwhile, a sharp increase in the expression of MDH protein was observed in the peel and pulp tissues of fully developed fruit. It is tempting to speculate that the MDH participates in the carbon fixation step that occurs during banana fruit development, which was previously reported in mango pulp, where MDH was found to be up-regulated in pre-climacteric stage compared to climacteric stage [49].
4.6. PPIs of Identified Proteins
In a living cell, protein establishes a diverse functional connection with each other which are underlying in the cellular process [50]. Enolase, which catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate, remains the central core protein in the interacting network. The involvement of enolase in the energy metabolism and carbon flow process through glycolysis pathway was reported in previous studies during fruit ripening [35,49,51]. During kiwi fruit ripening, enolase is suggested to interact with other functional categories of proteins such as defense, i.e., pyruvate decarboxylase and protein storage (HSP 70) [51]. Previously, a strong induction of enolase by ethylene and chilling treatment was reported in kiwifruit and tomato, and suggested to be involved in climacteric fruit ripening [52]. Down-regulation of enolase enzymes in mango pulp during the ripening stage indicated a slower carbon flow through the glycolysis pathway [49]. In the current study, abundant expression of enolase during fruit ripening in the pulp tissue indicates that it may play a similar role in inducing the ethylene biosynthesis process in bananas during fruit ripening (Fig. 10).
In the present study, enolase protein was present in 40-DAF peel and pulp, 60-DAF pulp, 90-DAF peel, 6-DAR pulp, and 12-DAR pulp. However, the abundance of enolase protein was found to be high in 6-DAR pulp, suggesting an increase in the carbon flow through the glycolysis pathway is promoting banana fruit ripening.
PGM serves as an intermediate, which catalyzes the readily reversible inter-conversion of G-1P and glucose-6-phosphate during the glycolysis pathway [22]. PGM proteins have a pivotal role in energy metabolism and are significantly up-regulated during Chinese bayberry ripening [53]. PGM is involved in the sugar metabolism process in melons and olives during ripening, and the expression of PGM was relatively high in melons during ripening [22,54].
PGM acts as an important regulatory enzyme in cellular glucose utilization and energy homeostasis processes during orange ripening [55]. The down-regulated expression of PGM, together with the reduced carbon flow through the glycolysis pathway was reported in mangoes during ripening and in strawberries during development [12,49].
In the present study, PGM protein was present in the 40-DAF pulp, 6-DAR pulp, and 12-DAR peel. However, the abundance of PGM protein was significantly high at 6-DAR pulp, when compared to other stages. Our findings revealed that PGM plays a pivotal role in banana fruit ripening.
5. CONCLUSION
To our knowledge, this is the first in-depth proteome study that is focused on spatiotemporal expression of protein dynamics involved in different metabolic pathways in banana fruit. The current investigation on the spatiotemporal expression of proteins in the peel and pulp tissue discloses comprehensive information and sheds light on the key protein expression, tissue type, and its involvement in different metabolic pathways of banana fruit during development and ripening. A high abundance of ethylene biosynthesis-related protein namely SAM synthase was observed in 12-DAR peel during ripening, suggesting that the ethylene may play an active role in fruit ripening. Proteins involved in hormone regulation such as ARF-GAP and TPR_domain containing protein related to auxin and ABA biosynthesis exhibited high abundance during banana ripening, suggesting that the auxin and ABA may have a key role in promoting fruit ripening, other than ethylene in banana. XTH which is considered to be a major cell wall modification protein, displayed an increase in the protein expression in peel tissue during the later stage of banana ripening, i.e., 12-DAR peel, which suggests that the XTH proteins actively participate in the regulation of cell wall modification process during peel ripening. Further, some of the unique or distinct proteins were identified in specific stages of banana during development and ripening. PPI revealed that two glycolysis pathway-related proteins namely, PGM and enolase found to be interacting more with other protein partners in the network. The result from our study not only provides an insight into the protein changes during banana development and ripening process, but also enlightens the path for further utilization of these identified proteins for banana crop improvement programs. Through genome editing or recent biotechnological approaches functional role of the identified proteins can be assigned and utilized further in controlling the ripening process, thereby minimizing the post-harvest losses in bananas due to the over ripening.
6. ACKNOWLEDGMENTS
This study was supported by the research grant to G.K. Surabhi by Rastriya Krishi Vikash Yojana, Government of India (No. AG (RKVY) 04/2017-9975/Ag.dt.22.06.2017; OR/RKVY-HORT/2017/774), Science & Technology Department, Government of Odisha (No.27552800232014/202830, STBBSR, dt.17.7.2015) and Forest, Environment and Climate Change Department, Government of Odisha, India, is gratefully acknowledged. The authors wish to acknowledge the Mass Spectrometry Facility at IIT Bombay (MASSFIITB) supported by the Department of Biotechnology (BT/PR13114/INF/22/206/2015) for their support with the mass spectrometry analysis of the samples. The authors wish to thank the Chief Executive, Regional Plant Resource Centre for extending the facilities.
7. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
8. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
11. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
12. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Bhuiyan FR, Campos NA, Swennen R, Carpentier S. Characterizing fruit ripening in plantain and Cavendish bananas: a proteomics approach. J Proteom 2020;214:103632; CrossRef
2. Wolf S, Mouille G, Pelloux J. Homogalacturonan methyl-esterification and plant development. Mol Plant 2009;2:851–60; CrossRef
3. Jiang B, Ou S, Xu L, Mai W, Ye M, Gu H, et al. Comparative proteomic analysis provides novel insights into the regulation mechanism underlying papaya (Carica papaya L.) exocarp during fruit ripening process. BMC Plant Biol 2019;19(1):1–13; CrossRef
4. Lindo-García V, Giné-Bordonaba J, Leclerc C, Ubach D, Larrigaudiere C. The relationship between ethylene-and oxidative-related markers at harvest with the susceptibility of pears to develop superficial scald. Postharvest Biol Technol 2020;163:111135; CrossRef
5. Wang X, Pan L, Wang Y, Meng J, Deng L, Niu L, et al. PpIAA1 and PpERF4 form a positive feedback loop to regulate peach fruit ripening by integrating auxin and ethylene signals. Plant Sci 2021;313:111084; CrossRef
6. Gupta K, Wani HS, Razzaq A, Skalicky M, Samantara K, Gupta S, et al. Abscisic acid: role in fruit development and ripening. Front Plant Sci 2022;13:817500; CrossRef
7. Inaba A, Liu X, Yokotani N, Yamane M, Lu WJ, Nakano R, et al. Differential feedback regulation of ethylene biosynthesis in pulp and peel tissues of banana fruit. J Exp Bot 2007;58(5):1047–57; CrossRef
8. Jiang L, Kang, R, Feng L, Yu Z, Luo H. iTRAQ-based quantitative proteomic analysis of peach fruit (Prunus persica L.) at different ripening and postharvest storage stages. Postharvest Biol Technol 2020;164:111137; CrossRef
9. Liu Z, Lv J, Liu Y, Wang J, Zhang Z, Chen W, et al. Comprehensive phosphoproteomic analysis of pepper fruit development provides insight into plant signalling transduction. Int J Mol Sci 2020;21(6):1962; CrossRef
10. Al Obaidi JR, Jamil NAM, Rahmad N, Rosli NHM. Proteomic andmetabolomic study of wax apple (Syzygium samarangense) fruit during ripening process. Electrophoresis 2018;39(23):2954–64; CrossRef
11. Kok SY, Namasivayam P, Ee GCL, Ong-Abdullah M. Comparative proteomic analysis of oil palm (Elaeis guineensis Jacq.) during early fruit development. J Proteom 2021;232:104052; CrossRef
12. Li L, Wu Q, Wang Y, Aghdam MS, Ban Z, Zhang X, et al. Systematically quantitative proteomics and metabolite profiles offer insight into fruit ripening behavior in Fragaria× ananassa. RSC Adv 2019;9(25):14093–108; CrossRef
13. Hu H, Liu Y, Shi GL, Liu YP, Wu RJ, Yang AZ, et al. Proteomic analysis of peach endocarp and mesocarp during early fruit development. Physiol Plant 2011;142(4):390–406; CrossRef
14. Ning T, Chen C, Yi G, Chen H, Liu Y, Fan Y, et al. Changes in homogalacturonan metabolism in banana peel during fruit development and ripening. Int J Mol Sci 2022;23(1):243; CrossRef
15. Shafiee R, Nahvi I, Emtiazi G. Bioconversion of raw starch to SCP by coculture of Cryptococcus aerius and Saccharomyces cerevisiae. J Biol Sci 2005;5(6):717–23.
16. Franscistt W, David FB, Robert MD. The estimation of the total soluble carbohydrate in cauliflower tissue. In: Witham FH, Blaydes DF, Devlin RM, (eds.). Experiment in plant phyisiology, Reinhold Camp, New York, NY, p 16, 1971.
17. Carpentier SC, Witters E, Laukens K, Deckers P, Swennen R, Panis B. Preparation of protein extracts from recalcitrant tissues: an evaluation of different methods for two-dimensional gel electrophoresis analysis, Proteomics 2005;5(10):2497–507; CrossRef
18. Ray JA, Kushnir MM, Bunker A, Rockwood AL, Meikle AW. Direct measurement of free oestradiol in human serum by equilibrium dialysis–liquid chromatography–tandem mass spectrometry and reference intervals of free estradiol in women. Clin Chim Acta 2012;413:1008–14; CrossRef
19. Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:214–20; CrossRef
20. Yun Z, Li T, Gao H, Zhu H, Gupta VK, Jiang Y, et al. Integrated transcriptomic, proteomic, and metabolomics analysis reveals peel ripening of harvested banana under natural condition. Biomolecules 2019;(9):167; CrossRef
21. Schmitz GJH, Freschi L, Ferrari RC, Peroni-Okita FHG, Cordenunsi-Lysenko BR. Exploring the significance of photosynthetic activity and carbohydrate metabolism in peel tissues during banana fruit ripening. Sci Hortic 2022;295:110811; CrossRef
22. Schemberger MO, Stroka MA, Reis L, Los KKDS, de Araujo GAT, Sfeir MZT, et al. Transcriptome profiling of non-climacteric ‘yellow’ melon during ripening: insights on sugar metabolism. BMC Genomics 2020;21:262; CrossRef
23. Chin CF, Teoh EY, Chee MJY, Al-Obaidi JR, Rahmad N, Lawson T. Comparative proteomic analysis on fruit ripening processes in two varieties of tropical mango (Mangifera indica). Protein J 2019;38(6):704–15; CrossRef
24. Umer MJ, Safdar LB, Gebremeskel H, Zhao S, Yuan P, Zhu H, et al. Identification of key gene networks controlling organic acid and sugar metabolism during watermelon fruit development by integrating metabolic phenotypes and gene expression. Hort Res 2020;7(1):1–13; CrossRef
25. Tian X, Zhu LL, Yang N, Song J, Zhao H, Zhang J, et al. Proteomics and metabolomics reveal the regulatory pathways of ripening and quality in post-harvest kiwifruits. J Agric Food Chem 2021;69(2):824–35; CrossRef
26. Li X, Li C, Sun J, Jackson A. Dynamic changes of enzymes involved in sugar and organic acid level modification during blueberry fruit maturation. Food Chem 2019;309:125617; CrossRef
27. Iqbal N, Khan NA, Ferrante A, Trivellini A, Francini A, Khan MI. Ethylene role in plant growth, development and senescence: interaction with other phytohormones. Front Plant Sci 2017;8:475; CrossRef
28. Wang S, Liu J, Zhao T, Du C, Nie S, Zhang Y, et al. Modification of Threonine-1050 of SlBRI1 regulates BR signaling and increases fruit yield of tomato. BMC Plant Biol 2019;19(1):1–13; CrossRef
29. Choi HR, Baek MW, Cheol LH, Jeong CS, Tilahun S. Changes in metabolites and antioxidant activities of green ‘Hayward’ and gold ‘Haegeum’ kiwifruits during ripening with ethylene treatment. Food Chem 2022;384:132490; CrossRef
30. Faurobert M, Mihr C, Bertin N, Pawlowski T, Negroni L, Sommerer N, et al. Major proteome variations associated with cherry tomato pericarp development and ripening. Plant Phys 2007;143(3):1327–46; CrossRef
31. Zhuang X, Jiang J, Li J, Ma Q, Xu Y, Xue Y, et al. Over-expression of OsAGAP, an ARF-GAP, interferes with auxin influx, vesicle trafficking and root development. Plant J 2006;48(4):581–91; CrossRef
32. Sieburth LE, Muday GK, King EJ, Benton G, Kim S, Metcalf KE, et al. SCARFACE encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. Plant Cell 2006;18(6):1396–411; CrossRef
33. Zemlyanskaya EV, Omelyanchuk NA, Ubogoeva EV, Mironova VV. Deciphering auxin-ethylene crosstalk at a systems level. Int J Mol Sci 2018;19(12):4060; CrossRef
34. Yoshida H, Nagata M, Saito K, Wang KL, Ecker JR. Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases, BMC Plant Biol 2005;5:14; CrossRef
35. Li T, Yun Z, Wu Q, Qu H, Duan X, Jiang Y. Combination of transcriptomic, proteomic, and metabolomic analysis reveals the ripening mechanism of banana pulp. Biomolecules 2019;9(10):523; CrossRef
36. Xiao L, Li T, Jiang G, Jiang Y, Duan X. Cell wall proteome analysis of banana fruit softening using iTRAQ technology. J Proteom 2019;209:103506; CrossRef
37. Meli VS, Ghosh S, Prabha TN, Chakraborty N, Chakraborty S, Datta A. Enhancement of fruit shelf life by suppressing N-glycan processing enzymes. Proc Natl Acad Sci 2010;107(6):2413–8; CrossRef
38. Ghosh S, Meli VS, Kumar A, Thakur A, Chakraborty N, Chakraborty S, et al. The N-glycan processing enzymes α-mannosidase and β-D-N-acetyl hexosaminidase are involved in ripening-associated softening in the non-climacteric fruits of capsicum. J Exp Bot 2011;62(2):571–82; CrossRef
39. Zhao Y, Zhu X, Hou Y, Wang X, Li X. Effects of nitric oxide fumigation treatment on retarding cell wall degradation and delaying softening of winter jujube (Ziziphus jujuba Mill. cv. Dongzao) fruit during storage. Postharvest Biol Technol 2019;156:110954; CrossRef
40. Huan C, An X, Yu M, Jiang L, Ma R, Tu M, et al. Effect of combined heat and 1-MCP treatment on the quality and antioxidant level of peach fruit during storage. Postharvest Biol Technol 2018;145:193–202; CrossRef
41. Niu J, Shi Y, Huang K, Zhong Y, Chen J, Sun Z, et al. Integrative transcriptome and proteome analyses provide new insights into different stages of Akebia trifoliata fruit cracking during ripening. Biotechnol Biofuels 2020;13(1):1–18; CrossRef
42. Fan Z, Kuang J, Fu C, Shan W, Han Y, Xiao Y, et al. The Banana transcriptional repressor MaDEAR1 negatively regulates cell wall-modifying genes involved in fruit ripening. Front Plant Sci 2016;7:1021; CrossRef
43. Chen S, Li Y, Zhao Y, Li G, Zhang W, Wu Y, et al. iTRAQ and RNA-Seq. analyses revealed the effects of grafting on fruit development and ripening of oriental melon (Cucumis melo L. var. makuwa). Gene 2021;766:145142; CrossRef
44. Kim J, Lee J, Hong Y, Lee EJ. Analysis of eight phytohormone concentrations, expression levels of ABA biosynthesis genes, and ripening-related transcription factors during fruit development in strawberry. J Plant Physiol 2019;239:52–60; CrossRef
45. Phillips HR, Landis JB, Specht CD. Revisiting floral fusion: the evolution and molecular basis of a developmental innovation. J Exp Bot 2020;71(12):3390–404; CrossRef
46. Bianco L, Alagna F, Baldoni L, Finnie C, Svensson B, Perrotta G. Proteome regulation during Olea europaea fruit development. PLoS One 2013;8(1)1–18; CrossRef
47. Yu JQ, Gu KD, Sun CH, Zhang QY, Wang JH, Ma FF, et al. The apple bHLH transcription factor MdbHLH3 functions in determining the fruit carbohydrates and malate. Plant Biotech J 2020;19(2):285–99; CrossRef
48. Zhang G, Cui X, Niu J, Ma F, Li P. Visible light regulates anthocyanin synthesis via malate dehydrogenases and the ethylene signalling pathway in plum (Prunus salicina L.), Physiol Plant 2021;172(3):1739–49; CrossRef
49. de Magalhaes Andrade J, Toledo TT, Nogueira SB, Cordenunsi BR, Lajolo FM, do Nascimento JRO. 2D-DIGE analysis of mango (Mangifera indica L.) fruit reveals major proteomic changes associated with ripening. J Proteom 2012;75(11):3331–41; CrossRef
50. Miernyk JA, Thelen JJ. Biochemical approaches for discovering protein-protein interactions. Plant J 2008;53:597–609; CrossRef
51. Minas IS, Tanou G, Krokida A, Karagiannis E, Belghazi M, Vasilakakis M, et al. Ozone-induced inhibition of kiwifruit ripening is amplified by 1-methylcyclopropene and reversed by exogenous ethylene. BMC Plant Biol 2018;18(1):1–19; CrossRef
52. Minas IS, Tanou G, Karagiannis E, Belghazi M, Molassiotis A. Coupling of physiological and proteomic analysis to understand the ethylene-and chilling-induced kiwifruit ripening syndrome. Front Plant Sci 2016;7:120; CrossRef
53. Chen YY, Zhang ZH, Zhong CY, Song XM, Lin QH, Huang CM, et al. Functional analysis of differentially expressed proteins in Chinese bayberry (Myrica rubra Sieb. et Zucc.) fruits during ripening. Food Chem 2016;190:763–70; CrossRef
54. Wu MC, Hu HT, Yang L, Yang L. Proteomic analysis of up-accumulated proteins associated with fruit quality during autumn olive (Elaeagnus umbellata) fruit ripening. J Agric Food Chem 2011;59(2):577–83; CrossRef
55. Zeng Y, Pan Z, Wang L, Ding Y, Xu Q, Xiao S, et al. Phosphoproteomic analysis of chromoplasts from sweet orange during fruit ripening. Physiol Plant 2014;150(2):252–70; CrossRef