1. INTRODUCTION
Traditional medical systems have been the primary healthcare approach in many countries for centuries. Despite the dominance of Western scientific medicine, many people are returning to traditional medical systems, substituting conventional therapies with traditional practices [1]. The accurate identification of plants used in the production of herbal medicines is essential for the safety of consumers.
Advances in genetics have facilitated species identification technologies such as DNA barcoding. The DNA barcoding technique uses short, standardized DNA segments for species identification, functioning like a unique fingerprint for each species. In 2003, Paul Hebert proposed using a short genetic sequence of <1,000 bp in length from the genome for species identification [2]. The mitochondrial gene cytochrome c oxidase I was established as the core for a global bio-identification system for animals, and the chloroplast DNA, containing variable regions such as maturase K (matK), ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcl), and Internal transcribed spacer (ITS) has been commonly used for DNA barcoding studies in plants [3]. In a recent study, the markers rbcL, matK, rbcL + matK, and psbA-trnH have also been utilized in the identification of 137 invasive plant species belonging to 11 families [4].
The matK gene, with its 1,500 bp length and high substitution rate, shows significant potential for resolving evolutionary and systematic issues across various taxonomic levels [5,6]. It meets the criteria for an effective DNA barcode, including significant species-level variability, conserved flanking regions for universal PCR primers, and suitability for bioinformatic analysis. When morphological identification is challenging, DNA barcoding can identify species from trace tissues, leaves, seeds, sterile seedlings, or fragmentary materials [7]. This approach can accurately differentiate even closely related species that appear similar in morphology, guaranteeing precise identification. The matK alone was sufficient to differentiate the species in Hibiscus with 100% resolution and 0.3% to 6.5% divergence [8]. Similarly, matK exhibited the highest interspecific divergence compared to the other markers, such as psbA-trnH, ITS2, and rbcL, in the identification of species in the genus Ardisia, identifying 22 out of 24 species [9]. This suggests that matK can be effectively used as a marker for the identification of subspecies in plants.
The current status of DNA barcoding for Indian medicinal plants reveals a mixed landscape, with significant progress made in certain areas but notable gaps remaining. While numerous species have been successfully barcoded, providing a robust tool for authentication and conservation, some important medicinal plants still need comprehensive barcode data [10]. Some plant species have many varieties and have indicated some valuable medicinal values [11–14]. However, it is difficult to identify them because of their similar morphological characteristics. Meanwhile, efforts have been made to generate DNA barcodes for genera such as Nerium (Oleander), Catharanthus, Cassia, and many more [15–17].
Bougainvillea, from the family Nyctaginaceae, is known for its vivid, colorful bracts, often mistaken for flowers. These bracts are modified leaves, 1/2–2 inches long, with actual flowers attached at the mid-rib [18]. Primary varieties include Bougainvillea spectabilis, Bougainvillea glabra, Bougainvillea peruviana, and their hybrids and cultivars. The bract color in B. glabra comes from betalain pigments [19]. Traditionally, it is used to treat respiratory conditions such as cough, asthma, bronchitis, and gastrointestinal disorders. It also has antibacterial and antifungal properties [19]. While another such plant, Dianthus, a member of the Caryophyllaceae family, contains varieties such as D. Chinensis, D. grantianopolitanus, D. barbatus, D. japonicus, and many more. It is used in traditional Chinese medicine to treat urinary tract infections, wounds, and skin inflammations. Ethnomedicinally, it is used for menostasis and gonorrhea, as a diuretic, emmenagogue, and coughs [20–22]. Similarly, Plumeria, commonly known as frangipani, belongs to the family Apocynaceae. These tropical flowering plants are renowned for their fragrant and beautiful flowers. Primary varieties include Plumeria obtusa, Plumeria rubra, Plumeria alba, and Plumeria cubensis [23]. Plumeria species exhibit potent antimicrobial properties against E. coli, potentially serving as a non-toxic antibiotic source. It treats malaria, leprosy, and skin ailments such as herpes and ulcers. Its bark is applied to tumors, while seeds have hemostatic properties [24]. These commonly occurring species have huge potential for medicinal uses. However, it is challenging to identify the correct species due to their very similar morphological characteristics.
In this study, we have used matK as a potential marker to differentiate species among medicinal plants such as Bougainvillea, Dianthus, and Plumeria, which are less represented in the DNA barcode library. The sequence analysis showed that matK genes for these species have notable polymorphism, which can be used to differentiate among the species. The phylogenetic analysis revealed that our matK has clustered with similar related species in the Bougainvillea, Dianthus, and Plumeria genera. We have also generated two-dimensional DNA QR codes that can be scanned easily.
2. MATERIALS AND METHODS
2.1. Plant Material Collection and DNA Extraction
The fresh leaves of three inter- or intra-species variants of Bougainvillea, Dianthus, and Plumeria were collected from the Indian Institute of Technology Gandhinagar (IITGN) campus in India. The images were captured using a camera. The details for the species variants and their morphological characters are provided in Supplementary Table 1. Genomic DNA was extracted from approximately 50–100 mg of fresh leaf samples using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA) following the manufacturer’s protocol. The concentration and purity of the extracted DNA were determined using a Nanodrop spectrophotometer.
2.2. Primer Designing, PCR Amplification and Sequencing
The primers for the Bougainvillea, Dianthus, and Plumeria matK genes were designed using Primer3 (https://primer3.ut.ee/), and the specificity of the primers was analyzed using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). For designing the primers, the matK gene sequences (Five to six in number) for the respective genera were collected from NCBI, and multiple sequence alignment was performed using Clustal Omega (https://www.ebi.ac.uk/jdispatcher/msa/clustalo). The primers were designed from the highly conserved regions determined using multiple sequence alignment with the amplicon size of 500–700 nucleotide (nt) and comprised of regions with the polymorphism. The universal primers M13F and M13R sites were added to the designed primer sequences to facilitate the sequencing. The details for the primers are given in Table 1.
PCR (20 µl) consisted of 10 μl EmeraldAmp® GT PCR Master Mix (Takara Bio, Japan), 0.4 µM of each forward and reverse primer, and 100–150 ng of DNA. The PCR conditions included initial denaturation for 5 minutes at 95°C, followed by 35 cycles of denaturation for 1 minute at 95°C, 45 seconds of annealing at 52°C–54°C, and 45 seconds extension, with a final extension of 5 minutes at 72°C. The PCR amplicon size was analyzed on the 1% agarose gel. The PCR amplicons were cleaned using QIAquick® PCR Purification Kit (Qiagen, Valencia, CA) following the manufacturer protocol and sent for sequencing to Eurofins India, Bangalore. The Sanger sequencing method was used to sequence PCR amplicons.
2.3. Sequence and Phylogenetic Analysis
The PCR amplicons sequenced using universal M13 F and M13 R primers were aligned for each genus, and the high-quality reads were only used for further analysis. Sequence for each species was used to search against the NCBI NR database using BLASTn. The polymorphism among the species within the genera and their variants was recorded.
 | Table 1. Primer used for DNA barcoding in this study. [Click here to view] |
The nucleotide sequences for the matK genes for B. glabra (JX495674.1), B. spectabilis (JN114741.1), B. spinosa (KY952357.1), P. alba (FJ754255.1), P. rubra (JQ586553.1), P. cubensis (DQ660536.1), P. obtusa (KJ012725.1), D. gratianopolitanus (MK926158.1), D. chinensis (KU722868.1), and D. superbus (KU722875.1) were retrieved from NCBI database and used for the phylogenetic analysis with sequenced matK genes from this study. The phylogenetic analysis of a total of 19 matK gene sequences was performed using MEGA11 [25]. The matK gene sequences were aligned using MUSCLE, and the phylogenetic tree was constructed using the Neighbor-Joining method with the Kimura two-parameter model and 1,000 bootstraps. The phylogenetic tree was visualized using Tree of Life (iTOL) (https://itol.embl.de/login.cgi).
2.4. Two-Dimensional DNA Barcoding
The two-dimensional images for QR codes for each species were generated to direct to the DNA barcode sequence for the matK for the respective species. The barcode images were converted into two-dimensional images with the help of a QR generator (https://www.qr-code-generator.com/).
3. RESULT AND DISCUSSION
3.1. Bougainvillea, D. chinensis, and Plumeria Have Distinguished Morphological Characters
The three species collected for Bougainvillea, D. chinensis, and Plumeria had distinct morphological characters from bract or petal size, shape, and leaf morphology. The Bougainvillea species in this study had bract color variations such as reddish orange (B. glabra “RO”), white (B. spectabilis “W”), and reddish pink (B. spectabilis “P”). Dianthus species exhibited color variations, such as white and pink (D. chinensis “WP”), pink and dark pink (D. chinensis “PDP”), and pure white (D. chinensis “W”). Similarly, Plumeria species showed variations in colors for petals such as white and yellow with overlapping petals (P. rubra cv. Acutifolia), red (P. rubra), and white and yellow with open and long petals (P. obtusa). The images for the bract or flowers of species used in this study are given in Figure 1. Some of the past studies have shown that B. glabra and B. spectabilis share a similar overall appearance, with the main differences being their bloom cycles and leaf characteristics; B. glabra has hairless, glossy leaves, while B. spectabilis features hairy leaves [18]. However, it is very challenging to differentiate these species using morphological characters.
3.2. Sequenced matK Regions had Notable Polymorphism Among Species
The PCR amplification of the matK gene produced clear, distinct bands of the single expected size on agarose gel, confirming the specificity of our primers (Fig. 2). The amplicon sizes for Bougainvillea, D. chinensis, and Plumeria were 681, 637, and 578 nt, respectively. The PCR amplicons sequenced using universal M13F and M13 R primers had good-quality sequence reads (Supplementary Table 2). The sequences were trimmed to 545–624 nt for barcode generations. The sequences were deposited to the NCBI database (Table 2).
The multiple sequence alignment for Bougainvillea, Dianthus, and Plumeria exhibited a notable polymorphism of approximately 1% to 2.7% among the inter and intra-species (Supplementary Fig. 1A–C). These results suggest that matK can be used as a potential marker for the successful differentiation of very close inter and intra-species. Nevertheless, it is crucial to design the primers from the conserved regions of the species in such a way that it will cover the highly polymorphic regions for the genetic marker, such as matK. Also, the addition of universal M13 F and M13 R sites to the primers can facilitate the sequencing of marker genes and avoid hassles in cloning PCR fragments.
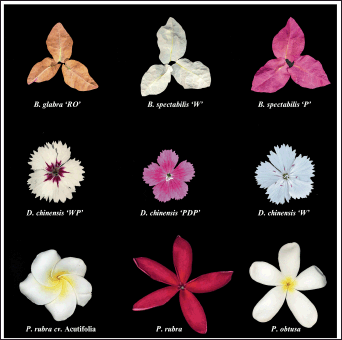 | Figure 1. Species used for DNA barcoding in this study. Three variants of Bougainvillea (B. glabra ‘RO’, B. spectabilis ‘W’, and B. spectabilis ‘P’) Dianthus (D. chinensis ‘WP’, ‘PDP’, ‘W’), and Plumeria (P. rubra cv. Acutifolia, P. rubra, and P. obtusa) used in this study are shown in the Figure. [Click here to view] |
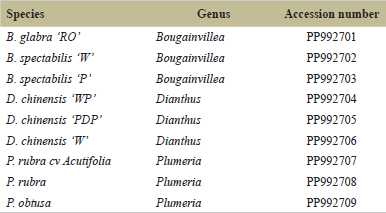 | Table 2. NCBI identifiers for DNA barcodes generated in this study. [Click here to view] |
3.3. Sequence Analysis Exhibited Similarities with matK in Closely Related Species
The Bougainvillea species matK sequences in this study shared the identity of 98.4% to 99.84% at the nucleotide level with other Bougainvillea species, such as B. glabra and B. spectabilis at the NCBI database Precisely, B. glabra “RO” shared maximum identity with B. glabra (KY952356.1), B. spectabilis “W” with B. spectabilis (JN114741.1), and B. spectabilis “P” with both B. glabra and B. spectabilis to the extent of 99.5% to 99.84%. Despite differing flower colors, the BLAST search results were consistent with the findings of previous studies, which highlight the morphological similarities and close chloroplast genome relationships between B. spectabilis and B. glabra [19, 26–27]. It was also observed that these sequences also share a similar identity to other Bougainvillea species genomes, such as B. peruviana or B. spinosa. The observed variations in our sequences not differentiating in BLASTn search could be due to the unavailability of comprehensive sequences in GenBank, where many entries might represent cultivars or higher similarities between the matK genes for these species, especially in the case of B. glabra and B. spectabilis. There could also be a possibility of sequencing errors complicating differentiation for intra-related species in the NCBI database. Furthermore, many Bougainvillea hybrid cultivars, such as B × spectro-glabra and B. × specto-peruviana, result in less differentiation among these species [28].
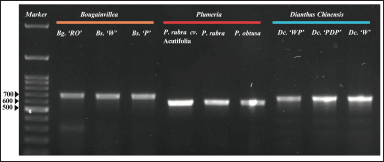 | Figure 2. Agarose gel electrophoresis of PCR amplified products for matK genes. The PCR-amplified products for matK Bougainvillea glabra (Bg. ‘RO’), Bougainvillea spectabilis (Bs ‘W’), and Bougainvillea spectabilis (Bs. ‘P’) denote the amplicon size of 681 n. Plumeria rubra cv. Acutifolia, Plumeria rubra, and Plumeria obtusa have the amplicon size of 578 nt. Dianthus chinensis (Dc. ‘WP’, ‘PDP’, and ‘W’) variants have an amplicon size of 637nt. [Click here to view] |
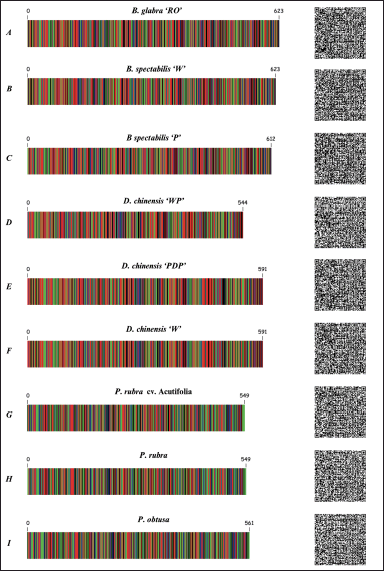 | Figure 3. Two-dimensional DNA barcodes generated for the species used in this study. The two-dimensional DNA barcodes for the Bougainvillea glabra (B. glabra ‘RO’, B. spectabilis ‘W’, and B. spectabilis ‘P’) Dianthus (D. chinensis ‘WP’, ‘PDP’, ‘W’), and Plumeria (P. rubra cv. Acutifolia, P. rubra, and P. obtusa) species were generated using QR generator (https://www.qr-code-generator.com/). DNA bar codes were generated using Bio-Rad DNA barcode generator (https://biorad-ads.com/DNABarcodeWeb/). [Click here to view] |
In the case of Dianthus, the sequences of D. chinensis in this study showed a maximum identity of 98.81% to 99.83% with the matK nucleotide sequences in the NCBI database. D. chinensis “WP” shared maximum identity with both D. grantianopolitanus and D. chinensis, while D. chinensis “PDP” and D. chinensis “W” exhibited identity with D. Chinensis. However, a similar percent identity for the matK was observed for various Dianthus species, indicating the close similarity in this genus. The NCBI database did not have specific entries for the different color variants of D. chinensis. This reflects a gap in botanical databases where phenotypic color variations are not always genetically differentiated or recognized as separate entries. The matK barcodes used efficiently detected the variations in our D. chinensis variants.
The BLASTn search for our Plumeria matK exhibited a clear differentiation among the species. Plumeria rubra, P. rubra cultivar Acutifolia (P. alba), and P. obtusa were easily distinguishable using the matK gene, demonstrating the marker’s robustness for this genus. However, it shared a similarity of 99.24% to 99.81% with Plumeria species matK nucleotide sequences in the NCBI database. The number of Plumeria species available ranges from 5 to 45, with P. rubra serving as the primary source for many Plumeria cultivars. Plumeria rubra is responsible for numerous color variations and many named cultivars available today. Historically, the different flower colors of P. rubra were considered separate species but are now recognized as variations within the same species. For instance, the P. rubra cultivar Acutifolia, which features white flowers with yellow centers, is often referred to as P. alba in some regions, as noted in a previous study conducted in Florida [29]. Consequently, P. alba has become a convenient term for the white-flowered form, encompassing numerous cultivars. The polymorphism in the matK for these species provided the robustness of this gene as a marker to differentiate these species and form barcodes, aligning with previous research on the efficacy of matK for Plumeria barcoding [23]. However, with all these limitations of sequence similarity with other genomes, erroneous deposited sequences, and hybrid cultivar genomes, the matK marker effectively differentiated the intra and inter-species we used in this study.
3.4. Phylogenetic Analysis Revealed the Clustering of Closely Related Species
The phylogenetic analysis of matK genes in this study with other ten matK partial nucleotide sequences retrieved from the NCBI database inferred using the neighbor-joining for the Bougainvillea, Dianthus, and Plumeria species showed that the closely related species in these genera clustered together. The Bougainvillea clade distinctly separates B. glabra (JX495674.1), Bougainvillea spinosa (KY952357.1), and B. spectabilis (JN114741.1). The newly sequenced matK for B. spectabilis “P” and B. glabra “RO”, align closely with B.glabra ((JX495674.1) reflecting their close morphological characteristics. Interestingly, B. spectabilis “P” was closer to B. glabra “RO”, indicating close similarity between the B. glabra and B. spectabilis genomes. The positioning of B. glabra “RO” within this clade suggests potential hybrid origins, supported by prior studies highlighting morphological similarities between B. glabra and B. spectabilis. Another possible explanation for this could be the higher level of sequence similarity between the matK of B. spectabilis and B. glabra, which makes it challenging to differentiate these species. This observation contradicts the principle that interspecific differences should be more pronounced than intraspecific variations, as suggested by the Consortium for the Barcode of Life [30]. Morphologically, it is visibly seen that B. glabra and B. spectabilis share a similar overall appearance, with the main differences being their bloom cycles and leaf characteristics; B. glabra has hairless, glossy leaves, while B. spectabilis features hairy leaves [18, 31,32]. Considering the origin of cultivars and the results from our phylogenetic analysis, it is plausible that previously published cp genome data for B. spectabilis may contain inaccuracies, potentially classifying it within the polyphyletic group of B. glabra or as a hybrid of B. glabra × B. spectabilis. The results for Bougainvillea align with previous studies, suggesting a high likelihood of errors in the existing Bougainvillea species database [32].
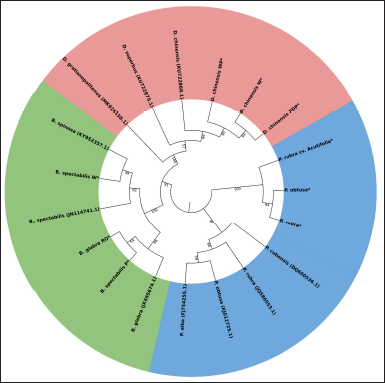 | Figure 4. Phylogenetic analysis of matK genes. The phylogenetic analysis for matK gene sequences collected for Bougainvillea, Plumeria, and Dianthus species, and matK genes sequenced in this study (represented by *) was carried out using the neighbor-joining method and Imura 2-parameter (K2P) model with 1000 bootstrap in MEGA11. The Phylogenetic tree is visualized using iTOL (https://itol.embl.de/). The nucleotide sequences for matK used in the DNA barcode generation in this study are denoted by Asterix(*). [Click here to view] |
The D. chinensis group showed clear differentiation among the sequenced samples, clustering with D. chinensis (KU722868.1). D. grantianopolitanus (MK926158.1) and D. superbus (KU722875.1) were well-separated from the D. chinensis cluster, indicating significant genetic differences among these species. Within the D. chinensis subgroup, the newly sequenced samples, labeled as D. chinensis “WP”, D. chinensis “PDP”, and D. chinensis “W”, formed a cohesive group, demonstrating their close genetic relationship. Notably, D. gratianopolitanus and D. superbus (KU722875.1) formed a distinct branch while D. chinensis matK clustered together, further underscoring the genetic distinction between these closely related species.
Interestingly, previously published sequences of P. alba and P. obtusa clustered together and formed a sister group with P. rubra. Simultaneously, P. rubra, P. rubra cv Acutifolia, and P. obtusa fell under the same sub-clade. Another surprising observation is that P. rubra, P. rubra cv Acutifolia, and P. rubra (JQ586553.1) did not cluster closely despite belonging to the same species. The possible reason for this would be that multiple studies have classified various Plumeria samples under a single scientific name despite different phenotypic characteristics, leading to ambiguity in the current databases. However, we have also observed the errors in the nomenclature of closely related species in the NCBI database. Either this may be due to an error in the identification of species due to similar morphology or genome similarity between the closely related species. These observations highlight the need for more precise and accurate identification of species and critical analysis of sequences.
The phylogenetic analysis highlighted both the effectiveness and limitations of the matK gene for DNA barcoding and species differentiation within Bougainvillea, Dianthus chinensis, and Plumeria, forming three distinct clades (Fig. 4). However, while matK shows substantial capability in differentiating genus and species, its utility for intraspecies identification has limitations and requires the correct species identification and annotations in the genomic databases.
3.5. QR Code for Barcodes can be Easily Scanned
The traditional two-dimensional DNA barcoding primarily includes the Latin names and sequence data of species. It would be beneficial to incorporate images of medicinal herbs and descriptions of their properties to enhance identification. This study transformed matK sequences into QR codes and two-dimensional DNA barcoding images (Fig. 3A–I). The colored DNA image uses various colors to represent different nucleotides, with numbers indicating sequence lengths, providing clear visual information [33]. Scanning the two-dimensional QR code next to the DNA barcodes (such as a mobile device) quickly provides the sequence for the barcodes.
3.6. Limitations and Challenges in Differentiation of Closely Related Species
The challenges involved in the identification of subspecies in the plants include the unavailability of the complete reference genome sequence, high sequence similarity for the marker genes among the subspecies, and sequencing quality and accuracy in the annotation of marker gene sequences in the databases. In the present study, the molecular marker matK demonstrated a robust differentiation capacity for the selected inter-intra species, effectively addressing practical identification issues. The matK was successfully used in the identification of subspecies of the Hibiscus and had similar resolutions to the other marker combinations, suggesting the robustness of the matK marker [8]. One of the factors in the success of the amplification of the matK or any other marker gene is the availability of high-quality sequence reads and correct annotation in the genome databases. The availability of full-length gene sequences for matK in databases such as NCBI for different species will be instrumental in designing primers in the flanking regions covering the regions with higher polymorphism among the species. Nevertheless, it should be noted that the use of a single matK as a marker may only work for some species, creating challenges in the resolution of closely related species. In such cases, another single marker can be used. For example, rbcL exhibited a higher differentiation potential than matK or a combination of matK and rbcL in the identification of jewel orchid accessions [34].
The closely related species in various plants have been identified using a combination of different markers such as matK, rbcL, ITS, ITS2, ndhF, ycf1, and psbA-trnH [35–39]. The previous studies have also suggested that the combination of regions such as trnH-psbA, trnL intron, trnL-trnF, psbI-trnG, and petA-psbJ could also enhance species differentiation [32]. The use of multiple combinations of such markers could be effective where the marker genes have higher sequence similarity among the subspecies. Overall, matK can be effectively used for the differentiation of closely related subspecies in some of the plants; however, for other species, a combination of markers along with matK can be used.
4. CONCLUSION
We have developed the matK as a genetic marker for the DNA barcoding of inter and intra-related species in the Bougainvillea, Dianthus, and Plumeria. The notable polymorphism for matK locus among a total of nine collected plant species variants studies demonstrates the overall potential of the matK gene for accurate species identification. The matK nucleotide sequences were submitted to the NCBI database to address the research gap. The matK sequences revealed significant genetic variability, providing a reliable tool for distinguishing species and variants used in this study. This study marks a positive step towards expanding the genetic barcode library and underscores the need for more comprehensive databases to improve species identification accuracy in medicinal plants. This research also provides a valuable reference for identifying plants with close phenotypic properties using DNA barcoding.
5. ACKNOWLEDGMENT
The authors sincerely acknowledge the IITGN for the Summer Research Internship Program to NP and W.O.O, postdoctoral fellowship to NG, SERB JRF to KHN, and fellowship to SP. The authors also acknowledge DBT for the Ramalingaswami Re-entry fellowship grant, SERB, and the IITGN for a start-up grant to SS.
6. AUTHOR CONTRIBUTION
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
7. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. DATA AVAILABILITY
The sequences for matK genes for nine inter and intra-species variants are deposited at NCBI. The accession numbers for the species include B. glabra ‘RO’ (PP992701), B. spectabilis ‘W’ (PP992702), B. spectabilis ‘P’ (PP992703), D. chinensis ‘W’ (PP992704), D. chinensis ‘PDP’(PP992705), D. chinensis ‘W’(PP992706), P. rubracv.Acutifolia (PP992707), P. rubra (PP992708), and P. obtusa (PP992709).
10. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
11. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
12. SUPPLEMENTARY MATERIAL:
The supplementary material can be accessed at the journal’s website: link here [https://jabonline.in/admin/php/uploadss/1282_pdf.pdf]
REFERENCES
1. Firenzuoli F, Gori L. Herbal medicine today: clinical and research issues. Evid Based Complement Alternat Med 2007;4(S1):594728; CrossRef
2. Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci 2003;270(1512):313–21; CrossRef
3. Pennisi E. Taxonomy. Wanted: a barcode for plants. Science 2007;318:190–1; CrossRef
4. Lonare N, Patil G, Waghmare S, Bhor R, Hardikar H, Tembe S. DNA barcoding of invasive terrestrial plant species in India. Mol Biotechnol 2024; CrossRef
5. Kress WJ, Erickson DL. DNA barcodes: genes, genomics, and bioinformatics. Proc Natl Acad Sci U S A 2008;105(8):2761–2; CrossRef
6. Hilu KW, Liang G. The matK gene: sequence variation and application in plant systematics. Am J Bot 1997;84(6):830–9; CrossRef
7. Von Cräutlein M, Korpelainen H, Pietiläinen M, Rikkinen, J. DNA barcoding: a tool for improved taxon identification and detection of species diversity. Biodivers Conserv 2011;20:373–89; CrossRef
8. Poovitha S, Stalin N, Balaji R, Parani M. Multi-locus DNA barcoding identifies matK as a suitable marker for species identification in Hibiscus L. Genome 2016;59(12):1150–6; CrossRef
9. Liu Y, Wang K, Liu Z, Luo K, Chen S, Chen K. Identification of medical plants of 24 Ardisia species from China using the matK genetic marker. Pharmacogn Mag 2013;9(36):331–7; CrossRef
10. Ekrem T, Willassen E, Stur E. A comprehensive DNA sequence library is essential for identification with DNA barcodes. Mol Phylogenet Evol 2007;43(2):530–42; CrossRef
11. Grover JK, Yadav S, Vats V. Medicinal plants of India with anti-diabetic potential. J Ethnopharmacol 2002;81(1):81–100; CrossRef
12. Zaheer Z, Konale AG, Patel KA, Khan S, Ahmed RZ. Comparative phytochemical screening of flowers of Plumeria alba and Plumeria rubra, Asian J Pharmaceutical Clinic Res 2010;3:88–9.
13. Chandra S, Rawat DS. Medicinal plants of the family Caryophyllaceae: a review of ethno-medicinal uses and pharmacological properties. Integr Med Res 2015;4(3):123–31; CrossRef
14. Ornelas García IG, Guerrero Barrera AL, Avelar González FJ, Chávez Vela NA, Gutiérrez Montiel D. Bougainvillea glabra Choisy (Nyctinaginacea): review of phytochemistry and antimicrobial potential. Front Chem 2023;11:1276514; CrossRef
15. Mahadani P, Sharma GD, Ghosh SK. Identification of ethnomedicinal plants (Rauvolfioideae: Apocynaceae) through DNA barcoding from northeast India. Pharmacogn Mag 2013;9(35):255–63; CrossRef
16. Purushothaman N, Newmaster SG, Ragupathy S, Stalin N, Suresh D, Arunraj DR, et al. A tiered barcode authentication tool to differentiate medicinal Cassia species in India. Genet Mol Res GMR 2014;13(2):2959–68; CrossRef
17. Arulvendhan V, Saravana Bhavan P, Rajaganesh R. Molecular identification and phytochemical analysis and bioactivity sssessment of Catharanthus roseus leaf extract: exploring a ntioxidant potential and antimicrobial activities. Appl Biochem Biotechnol 2024; CrossRef
18. Kobayashi K, Mcconnell J, Griffis Jr JL. Bougainvillea. Ornam Flowers 2007;38:1–12.
19. Abarca-Vargas R, Petricevich VL. Bougainvillea genus: a review on phytochemistry, pharmacology, and toxicology. Evid Based Complement Alternat Med 2018;2018:9070927; CrossRef
20. Lin S, Liu J, He X, Wang J, Wang Z, Zhang X, et al. Comprehensive comparative analysis and development of molecular markers for Dianthus species based on complete chloroplast genome sequences. Int J Mol Sci 2022;23(20):12567; CrossRef
21. Liu Q, Zang EH, Wang CC, Liu YC, Niu H, Gao Y, et al. Dianthi herba: a comprehensive review of its botany, traditional use, phytochemistry, and pharmacology. Chin Med 2022;17(1):15; CrossRef
22. Sreelekshmi R, Siril EA. An efficient in vitro thin cell layer multiplication and enhanced saponin production of Dianthus chinensis L.: a cultivated Chinese medicinal crop. Ind Crops Prod 2023;193:116194; CrossRef
23. Zhao L, Yu X, Shen J, Xu X. Identification of three kinds of Plumeria flowers by DNA barcoding and HPLC specific chromatogram. J Pharm Anal 2018;8(3):176–80; CrossRef
24. Sura J, Dwivedi DrS, Dubey R. Pharmacological, phytochemical, and traditional uses of Plumeria alba LINN. an Indian medicinal plant. SPER J Anal Drug Regul Aff 2016;1:14–7; CrossRef
25. Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38(7):3022–7; CrossRef
26. Bautista MAC, Zheng Y, Hu Z, Deng Y, Chen T. Comparative analysis of complete chloroplast genome sequences of wild and cultivated Bougainvillea (Nyctaginaceae). Plants 2020;9(12):1671; CrossRef
27. Zhang H, Huang T, Zhou Q, Sheng Q, Zhu Z. Complete chloroplast genomes and phylogenetic relationships of Bougainvillea spectabilis and Bougainvillea glabra (Nyctaginaceae). Int J Mol Sci 2023;24(17):13044; CrossRef
28. Salam P, Bhargav V, Gupta D, Nimbolkar P. Evolution in Bougainvillea (Bougainvillea Commers.)—a review. J Appl Nat Sci 2017;9:1489–94; CrossRef
29. Brown SH. Identification of the four forms of Plumeria rubra. Proc Fla State Hort Soc 2008;121:406–7.
30. CBOL Plant Working Group, Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, et al. Proc Natl Acad Sci 2009;106(31):12794–7; CrossRef
31. Kumar PP, Janakiram T, Bhat KV, Jain R, Prasad KV, Prabhu, KV. Molecular characterization and cultivar identification in Bougainvillea spp. using SSR markers. Indian J Agric Sci 2014;84(9):1024–30; CrossRef
32. Lin X, Lee SY, Ni J, Zhang X, Hu X, Zou P, et al. Comparative analyses of chloroplast genome provide effective molecular markers for species and cultivar identification in Bougainvillea. Int J Mol Sci 2023;24(20):15138; CrossRef
33. Liu C, Shi L, Xu X, Li H, Xing H, Liang D, et al. DNA barcode goes two-dimensions: DNA QR code web server. PLoS One 2012;7(5):e35146; CrossRef
34. Ho VT, Tran TKP, Vu TTT, Widiarsih S. Comparison of matK and rbcL DNA barcodes for genetic classification of jewel orchid accessions in Vietnam. J Genet Eng Biotechnol 2021;19(1):93; CrossRef
35. Choudhary A, Shekhawat D, Pathania J, Sita K, Sharma S, Chawla A, et al. Exploring DNA barcode for accurate identification of threatened Aconitum L. species from Western Himalaya. Mol Biol Rep 2024;51(1):75; CrossRef
36. Jin L, Shi HY, Li T, Zhao N, Xu Y, Xiao TW, et al. A DNA barcode library for woody plants in tropical and subtropical China. Sci Data 2023;10(1):819; CrossRef
37. Nath S, VanSlambrouck JT, Yao JW, Gullapalli A, Razi F, Lu Y. DNA barcoding of terrestrial invasive plant species in Southwest Michigan. Plant Direct 2024;8(6):e615; CrossRef
38. Sevindik E, Korkom Y, Murathan ZT. Evaluating DNA barcoding using cpDNA matK and rbcL for species identification and phylogenetic analysis of Prunus armeniaca L. (Rosaceae) genotypes. Genet Resour Crop Evol 2024;71:1825–35; CrossRef
39. Li H, Xiao W, Tong T, Li Y, Zhang M, Lin X, et al. The specific DNA barcodes based on chloroplast genes for species identification of Orchidaceae plants. Sci Rep 2021;11(1):1424; CrossRef