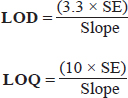1. INTRODUCTION
Malaria is a severe illness that poses a threat to human life and is caused by the Plasmodium parasite. Transmission of the disease occurs through the bite of female Anopheles mosquitoes [1]. The Plasmodium parasite represents a significant public health concern in sub-Saharan Africa, leading to nearly 1 million deaths annually. This high mortality rate is primarily attributed to the prevalence of Plasmodium falciparum, the most perilous human malaria parasite. Additionally, controlling malaria transmission in Africa is challenging due to the presence of the highly efficient malaria vector, Anopheles gambiae, which is widely distributed across the continent [2]. Over the last 50 years, P. falciparum has developed resistance to various antimalarial medications such as sulfadoxine/pyrimethamine, chloroquine, quinine, mefloquine, and piperaquine. Despite the advancements in rational drug design, natural products, particularly those derived from plants, remain a significant and valuable source of medicines nowadays [3].
Artemisia annua L. is a fragrant herb that contains artemisinin and essential oils. Artemisinin is an active antimalarial with less side effects than other antimalarials [4]. Artemisinin works by reacting with iron within the malaria parasite, producing free radicals that damage membranes and other vital structures within the parasite, leading to its death [5]. In 2006, the World Health Organization endorsed artemisinin combination treatments (ACTs) as the primary treatment option for both mild and severe cases of malaria [6]. In light of the emergence of resistance, artemisinin and its semisynthetic analogs, namely, artemether, arteether, and artesunate, are recognized as highly effective in treating uncomplicated malaria through ACTs and complex cerebral malaria through monotherapy [5]. In addition to antimalarial activity, artemisinin and its derivatives have a variety of additional bioactivities [7], including anticancer [8], antitumor [9], and antiviral [10].
Recently, the field of drug discovery and development has witnessed the growing prominence of computational bioinformatics due to its high efficiency. Computational approaches have become increasingly popular as they offer valuable tools for various stages of the drug research and development process. These methods have the potential to save significant time, financial resources, and human effort. Serine hydroxyl methyltransferase (SHMT) is an enzyme that plays a crucial role in the metabolism of P. falciparum, the parasite responsible for causing malaria in humans. This enzyme is involved in several metabolic pathways within the parasite, contributing to its survival and growth [11]. Due to its central role in folate metabolism, SHMT has been explored as a potential drug target for the treatment of malaria. Antifolate drugs, such as pyrimethamine and sulfadoxine, target enzymes involved in folate metabolism, including SHMT, to disrupt the parasite’s ability to synthesize DNA and RNA [12]. However, resistance to these drugs has emerged in some P. falciparum strains. This necessitates the rapid development of new class of inhibitors to antagonize the parasite folate metabolic pathways, thereby stopping and disrupting DNA and RNA synthesis in the parasite system. In this study, molecular docking and interaction studies were utilized, aiming to examine the binding affinity and molecular interactions between artemisinin and the SHMT enzyme of P. falciparum with PDB ID (1FDO). The objective was to identify the potential inhibitory activity of artemisinin against SHMT following the established method of Feng et al. [13] as the mechanism of action of artemisinin is yet to be revealed so far. Cold maceration technique (CMT) was also developed, which is safe, effective, low cost, and rapid for extracting artemisinin from A. annua plant. A validated HPTLC method was also established for the accurate and quick analysis of artemisinin using visible light detection at 550 nm. The results obtained from the four different extracts of A. annua were compared with that of standard and the extract with the highest quantity as well as high-quality (purity) artemisinin compound was finally screened. Therefore, an expedited sample preparation method and a validated procedure for the quantification of artemisinin utilizing the HPTLC method were established.
2. MATERIALS AND METHODS
2.1. Collection and Identification of Plant Material
A. annua L. plant with voucher number 8620-KASH was collected from Kawoosa Budgam, Kashmir, India, in August 2022. The collection site is located at coordinates (74.3051, 34.0614) and has an altitude of 1585 m. The plant’s authentication was conducted by a curator at Kashmir University, Department of Botany, Srinagar, Jammu and Kashmir, India. The authenticated plant specimens were deposited in the KASH herbarium. After collection, the plants were washed, subjected to shade drying for a period of 2–3 weeks until they were completely dry, and then powdered.
2.2. Chemicals and Reagents
Artemisinin (>99% purity) and HPLC-grade solvents, namely, methanol, chloroform, and petroleum ether, were purchased from Himedia, India. The chemicals, namely, anisaldehyde, glacial acetic acid, sulfuric acid, toluene, and acetone, were obtained from Sigma-Aldrich. Water purification was carried out using a Milli-Q water system manufactured by Millipore in Bedford, MA, USA.
2.3. Extraction by CMT
The powdered sample of A. annua L. was extracted using four different solvents: [petroleum ether → chloroform → methanol→ distilled water (ddH.O)]. About 5 g of plant samples was extracted in 20 ml of each solvent by cold maceration method starting from less polar to high polar solvents in the series [petroleum ether → chloroform → methanol→ distilled water (ddH.O)] successively. The powdered plant materials were kept in a conical flask in contact with the solvent in a stoppered container for a week with frequent agitation until the soluble matter was dissolved. The mixture of plant samples was allowed to soak for 3 days so that the plant molecules could move through the solvent according to their polarity. After 3 days, filtration was performed with Whatmann filter paper no. 1. The extracts were then concentrated using a rotary evaporator at a pressure of 600 mmHg (heating water set at a temperature of 50°C for chloroform and petroleum ether extracts and 60°C for methanol extract). For aqueous extracts, the evaporation of the solvents was achieved by keeping the extract in a hot air oven at 50°C at a pressure of 1 mmHg. The extracts generated were stored in sterile vials prior to HPTLC at 4°C.
2.4. Analysis of Artemisinin by HPTLC
2.4.1. Preparation of standard (Artemisinin)
To create a stock solution of artemisinin at a concentration of 1 mg/mL, 6 mg of artemisinin was dissolved in 6 mL of methanol. Subsequently, a final concentration of 0.1 mg/mL of artemisinin was achieved by dissolving 1 mL of the stock solution in 10 mL of methanol.
2.4.2. Preparation of test samples (Extracts)
To obtain a final concentration of 1 mg/mL, 10 mg of each extract was dissolved in 10 mL of their respective solvents. Subsequently, each sample solution was filtered using a 0.22 µm polypropylene membrane filter and then subjected to HPTLC analysis.
2.4.3. The setup conditions for HPTLC and instrumentation
All samples of four extracts were analyzed using the HPTLC system consisting of a Linomat-5 applicator CAMAG with a 100 µl syringe and a TLC scanner CAMAG (Switzerland) controlled using winCATS software (version 1.4.6.2002). HPTLC analysis was carried out on a precoated silica gel 60F254 TLC plate (10 × 10 cm). The mobile phase used for the analysis consisted of a mixture of petroleum ether and ethyl acetate at a ratio of 8.5:1.5 (75:25, v/v). For each sample of A. annua extracts, as well as the standard solution (artemisinin), 5 µl of each was applied to the plates as 6 mm bands. The concentration range of the standard solution applied varied from 5 to 20 µl. The sample application process was carried out using a Linomat-5 applicator from CAMAG, which was equipped with a 100-µl syringe. To ensure accuracy, the following settings were implemented: the band length was set to 6 mm, the application rate was set at 10 s/µl, a distance of 8 mm was maintained between the bands, and a distance of 1.5 cm was maintained from the side edge of the plate. The following variables were held constant for each analysis: (i) 75.0 mm developing distance; (ii) time to develop: 12–15 min; (iii) reagent for detection: anisaldehyde reagent; (iv) post-chromatographic derivatization time: 5 min in a hot air oven at 100°C; and (v) 550 nm densitometry scan. The anisaldehyde reagent, composed of 1 mL of anisaldehyde, 20 mL of glacial acetic acid, 170 mL of methanol, and 10 mL of concentrated sulfuric acid, was sprayed onto the plates. Subsequently, the plates were allowed to dry for 5 min at 100°C in a hot air oven. Following the formation of the bands, the plates underwent scanning using a Camag TLC scanner 3 at a reflectance wavelength of 550 nm. The resulting data were analyzed using winCATS software. The scanning condition for densitometry was 4.00 × 0.30 mm slit, with a scanning speed of 20 mm/s and a data resolution of 100 mm/step. The peak of the separated compounds from each extract was identified by comparing it to that of the standards, and the retention factor (Rf) value was determined. The concentration of each target compound was calculated using the calibration curves.
2.4.4. Linearity
The calibration curve of artemisinin was prepared by applying different volumes of standard solution (5, 10, 15, and 20 µl) to get the linearity range of 5–20 µg/spot. The calibration graph was created with peak area versus concentration at 550 nm wavelength. With the help of regression equation and consistent peak area, artemisinin yield was estimated.
2.4.5. Precision
To determine the precision and accuracy of the method, 300 ng/spot of artemisinin with (n = 4) was spotted in four different concentrations in triplicates. Reproducibility and repeatability of the results were determined by computing the coefficient of variation for intra-day and inter-day precision with different concentrations of artemisinin three times on the same day and after 3 days, respectively. These observations were made three times, and the results are expressed as mean ± %RSD.
2.4.6. Limit of detection (LOD) and limit of quantitation (LOQ)
The LOD and LOQ were calculated based on the standard error (SE) of the response and the slope of the calibration curve of quercetin using the following relations:
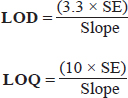
where SE = standard error of the response.
2.4.7. Densitometry and evaluation of peak spectra
To determine the purity and specificity of artemisinin from the extracts, densitometry scanning was performed using a CAMAG TLC scanner 3 in absorption mode. The scanner was connected to a personal computer running the user-friendly winCATS software (version 1.4.2). A deuterium lamp with a wavelength of 550 nm was chosen based on preliminary experiments as it corresponds to the near absorption maxima of the marker compounds. The scanning parameters include a slit dimension of 5 mm × 0.4 mm, a scanning speed of 20 mm/s, and a data resolution of 100 µm/step. The analysis involved evaluating artemisinin based on its retention factor (Rf) value, UV spectrum, and peak area measurement. The spectral wavelengths of the chromatographic peaks were monitored in the range of 400–550 nm. This methodology allowed for the precise characterization and quantification of artemisinin, providing valuable information about its presence, purity, and concentration in the tested extracts.
2.5. In Silico Inhibitory Activity of Artemisinin Against Serine Hydroxymethyl Transference (SHMT)
2.5.1. Protein and ligand preparation
The protein data bank https://www.rcsb.org/ was used to retrieve the 3D crystal structure of SHMT. However, before docking, the co-crystallized ligands, water molecules, and other heteroatoms were removed from the protein molecule using BIOVIA Discovery Studio 2021. All the ligand structures, including the control, were downloaded from the PubChem database [https://pubchem.ncbi.nlm.nih.gov/] and saved in SDF format. However, we also retrieved the structure of pyrimethamine. This ligand molecule was separately fetched from the PubChem database to serve as control in this study. A comparative study in terms docking score, ligand-binding mode, and stability through molecular interaction study and drug likeness was carried out. The ligands were all converted into PDBQT format prior to docking.
2.5.2. Virtual screening and molecular docking
To determine the binding affinity and other types of interactions between the artemisinin and the target protein, molecular docking was carried out using the PyRx tool, which is a virtual screening tool that employs Vina as well as Autodock 4.2. Previously prepared target protein and ligand molecule, including the control ligand molecule, were loaded in the PyRx virtual screening tool, the former being a macromolecule and the latter being ligands. A grid box was set around the receptor having dimensions of (X: 82.23, Y: 43.09, and Z: 77.52). The ligand molecules and the receptor molecule were docked, and the maximum exhaustiveness was computed for all the ligands. All other parameters in the software were kept in a default mode. The docking result was analyzed and compared with that of control. The protein–ligand binding interactions were profiled as per reported by Verma et al. [14] and Abhishek et al. [15].
2.6. Statistical Analysis
Experiments were replicated three times, and the values are expressed as mean ± standard deviation. The in silico results were analyzed by taking into account the binding affinity, suitable molecular interactions between the ligands and the receptor molecule, and ADMET analysis using standard bioinformatics tools as described in the methodology.
2.7. ADME Analysis
The ADME properties of the selected ligand were predicted using the SWISSADME website (http://www.swissadme.ch/index.php), which is an online tool that provides curated data for chemicals associated with known Absorption, Distribution, Metabolism, and Excretion (ADME) profiles [16].
2.8. Toxicity Analysis
Before the drug molecule was utilized as medicine, the toxicity analysis of the molecule should be done to ensure its safety. pkCSM (http://biosig.unimelb.edu.au/pkcsm/) is simply an online pharmacokinetics prediction database that predicts small molecules for ADMET properties. The molecule toxicity was analyzed by entering the ligand SMILE format into the database [17].
3. RESULTS AND DISCUSSION
3.1. Qualitative and Quantitative Estimation of Artemisinin by the HPTLC Densitometry Method
3.1.1. Development of optimum mobile phase ratio for artemisinin separation
The TLC mobile phase ratio was optimized to achieve proper separation of artemisinin. The poor separation of artemisinin was observed toward the end of the TLC plate while using solvent system of petroleum ether: ethyl acetate at a ratio of 8.5:1.5 (75:25, v/v) as shown in Figure 1. However, while adjusting the mobile ratio to ethyl acetate: petroleum ether at the ratio of (75:25, v/v), an excellent separation of artemisinin was observed toward the middle of the plate with a retention factor of (0.56) as shown in Figure 2. Therefore, the ethyl acetate:petroleum ether at the ratio of 8.5:1.5 (75:25, v/v) is a suitable solvent system for the extraction of artemisinin.
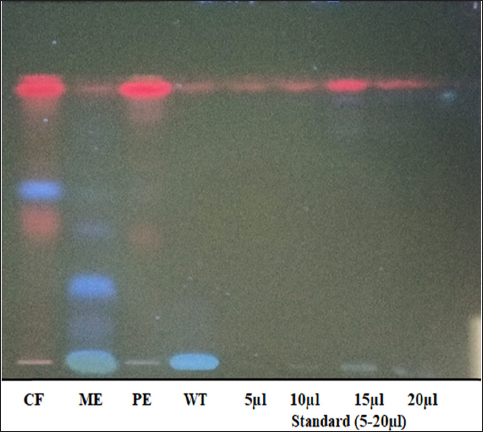 | Figure 1: HPTLC fingerprinting of four different extracts of Artemesia annua. CF: Chloroform extract. ME: Methanol extract.
PE: Petroleum ether extract. WT: Water extract. Artemisinin standard = (5–20 µl).
[Click here to view] |
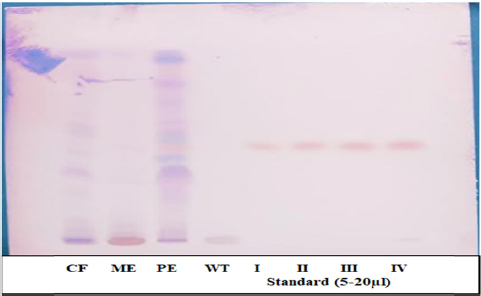 | Figure 2: HPTLC fingerprinting of four different extracts of Artemesia annua. CF: Chloroform extract. ME: Methanol extract.
PE: Petroleum ether extract. WT: Water extract. Accessions (I–1V) = artemisinin standard (5–20 µl).
[Click here to view] |
3.1.2. Influence of solvents on artemisinin yield
The selection of an appropriate solvent system is a crucial factor that significantly influences the separation of components on an HPTLC plate. This study aimed to isolate and quantify high-quality artemisinin compound from four different extracts of A. annua L. (whole plant) using high-performance thin-layer chromatography. To achieve this, four solvents with varying polarities were employed. The artemisinin content was determined by using the peak area parameter and percentage mean on a dry weight basis. The result obtained is presented in Table 1. Petroleum ether extract yields the maximum artemisinin content, followed by methanol and chloroform extract. However, very less amount of artemisinin was obtained in aqueous extract. Therefore, among all the studied extracts of A. annua, petroleum ether extract displayed a higher content of artemisinin compound (0.71% ± 0.02), followed by methanol extract (0.25% ± 0.02), chloroform extract (0.14% ± 0.01), and water extract (0.06% ± 0.01) as presented in Table 1 and Figure 3.
Table 1: Influence of solvents on artemisinin yield.
| S/no. | Extraction solvents | Mean % of Artemisinin ± SD
(dry wt. basis) | Rf value |
|---|
| 1 | Chloroform (CF) | 0.14 ± 0.01 | 0.59 |
| 2 | Methanol (ME) | 0.25 ± 0.02 | 0.56 |
| 3 | Petroleum ether | 0.71 ± 0.02 | 0.51 |
| 4 | Water (WT) | 0.06 ± 0.01 | 0.56 |
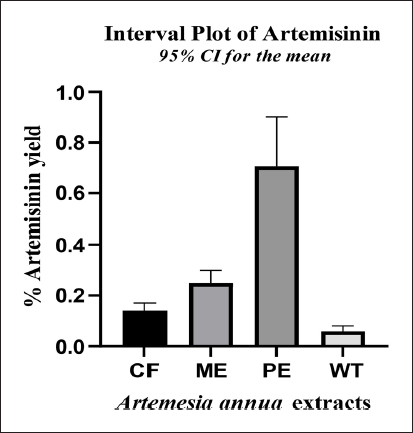 | Figure 3: Artemisinin content of different extracts of Artemisia annua plant. CF: Chloroform extract. ME: Methanol extract.
PE: Petroleum ether extract. WT: Water extract.
[Click here to view] |
A wide range of solvents have been used for artemisinin extraction, including hexane, ethanol, methanol, chloroform, petroleum ether, and ethyl acetate. Based on the previous study, ethanol was found to be very efficient for the artemisinin extraction. A study by Babacan et al. [18] found that ethanol was the most effective solvent for extracting artemisinin from A. annua. However, the present result shows the maximum yield of artemisinin in petroleum ether. Various studies have investigated the efficiency of petroleum ether in the extraction of artemisinin from A. annua. The studies by Cao et al. [19] and Kundu et al. [20] found that petroleum ether was able to extract artemisinin from A. annua, but the yield was lower compared to other solvents like designed hydrophobic and hydrophilic solvents. The study also found that the purity of artemisinin extracted using petroleum ether was higher than that obtained using other solvents. This is in agreement with the present finding as far as our densitometry graph is concerned [Figure 4]. Therefore, based on the previous study, petroleum ether was found to be less efficient in extracting artemisinin compared to other solvents like methanol, ethanol, and ethyl acetate. However, petroleum ether has been shown to selectively extract lipophilic compounds like artemisinic acid and casticin, which may have potential antimalarial activity. But the present study shows the efficiency of petroleum ether for extracting the maximum content as well as high-quality artemisinin with no impurities from A. annua.
 | Figure 4: Densitometry graph showing isolation of artemisinin compound from (A) chloroform extract, (B) methanol extract, (C) petroleum ether extract, (D) water extract of A. annua, and (E–H) artemisinin standard (5–20 µl) at 550 nm.
[Click here to view] |
3.1.3. Specificity
To confirm the specificity of the artemisinin compound in the extracts, a comparison was made between the Rf values of the extract and that of the standard bands as shown in Figure 4. The results indicate that the method employed was specific as it facilitated the clear separation of artemisinin compound in all four extracts.
3.1.4. Linearity
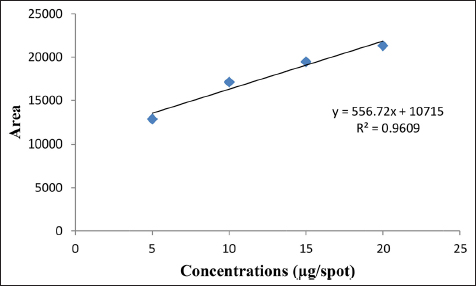
To assess the linearity, various concentrations of working standard solutions of artemisinin were applied. The resulting data were subjected to linear least-square regression analysis to determine the correlation coefficient (r2). A correlation coefficient (r2) of 0.9609 was obtained, indicating a robust linear association between the concentration of artemisinin and the corresponding response. The linearity range of the calibration curve for artemisinin was 5–20 µg/spot. Figure 5 illustrates the calibration curve graph, showcasing the linearity range of artemisinin.
3.2. Precision: Intra-Day and Inter-Day Precision
An intra-day and inter-day precision analysis method was carefully evaluated in four separate determinations of artemisinin (n = 4) on distinct tracks. Each track underwent scanning, and the resultant chromatograms were recorded. The consistency of these measurements was quantified using the percent coefficient of variation (RSD%) for the peak area of the artemisinin standard integrated into the extract. Intra-day precision was found to be 1.48% ± 0.001, and inter-day precision was found to be 1.50% ± 0.021. Figure 6 represents the 3D chromatogram data of both standard and extract while Figure 7 represents the spectral comparisons of artemisinin standard and that of the extracts. The results obtained affirmed that the method showcases a remarkable degree of precision for the analysis of artemisinin in A. annua plant. The presence of low RSD% values implies negligible variation in peak areas, underscoring the method’s exceptional measurement repeatability.
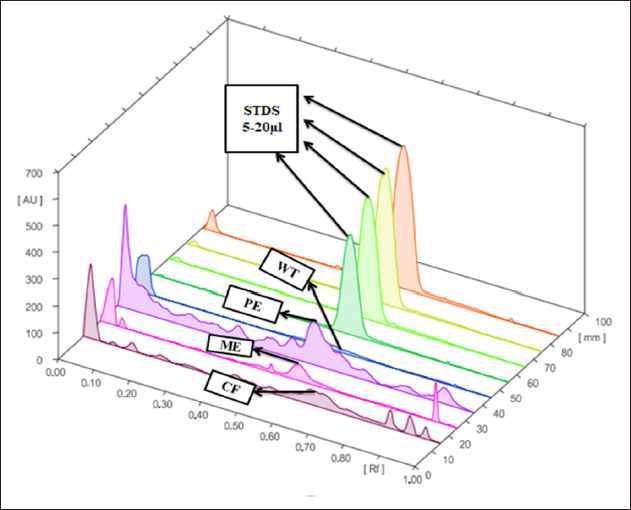 | Figure 6: 3D chromatogram of artemisinin from four different extracts of A. annua and standard (STDS). CF: Chloroform extract. ME: Methanol extract.
PE: Petroleum ether extract. WT: Water extract. STDS: artemisinin standard (5–20 µl).
[Click here to view] |
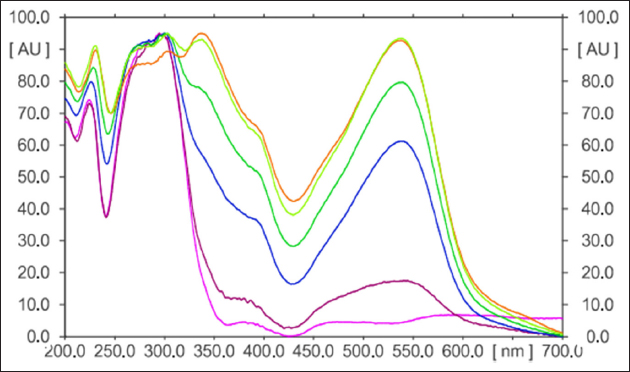 | Figure 7: Overlay spectral comparisons of the extracts and artemisinin standard.
[Click here to view] |
3.3. Limit of Detection and Limit of Quantitation
The limit of detection (LOD) of artemisinin was found to be 5.23 µg/spot while the limit of quantitation (LOQ) was found to be 15.93 µg/spot as shown in Table 5. The LOD and LOQ were calculated based on the SE of the response and slope as shown in Table 2. Therefore, this result demonstrates the method’s sensitivity and capability to detect and quantify artemisinin compound at low concentrations with a high degree of reliability and precision.
Table 2: HPTLC validated parameters.
| S/no. | Parameters | Artemisinin |
|---|
| 1 | Specificity | Specific |
| 2 | Regression equation | Y = 556.72X + 10715 |
| 3 | Correlation coefficient (r.) | 0.9609 |
| 4 | Correlation coefficient (r) | 0.9803 |
| 5 | Slope | 556.72 |
| 6 | Standard error of the slope | 882 |
| 7 | Linearity range (µg/spot) | 5-20 |
| 8 | Limit of detection (LOD) µg/spot | 5.23 |
| 9 | Limit of quantitation (LOQ) µg/spot | 15.93 |
3.4. Molecular Docking Analysis
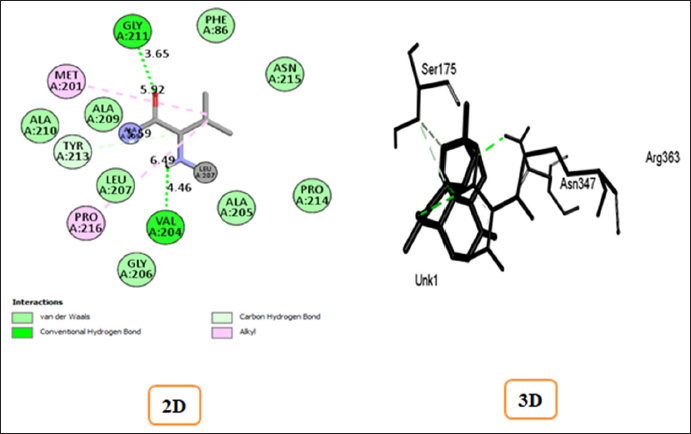
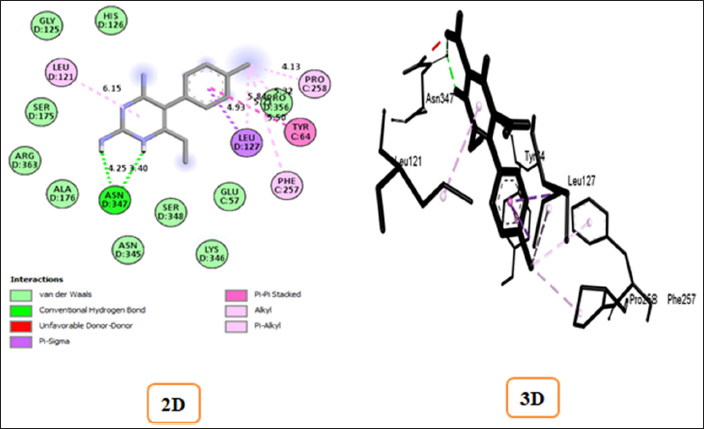
The ligand structures of artemisinin and pyrimethamine compounds obtained from the PubChem database were screened for their binding strength with the SHMT enzyme to determine binding energy and other affinities toward the active site. Figures 4 and 5 depict the complexes of artemisinin and pyrimethamine with SHMT in 2D and 3D formats, respectively. For artemisinin, the calculated binding affinity of −9.1 kcal/mol suggests a robust and favorable interaction with the SHMT enzyme. This high negative value indicates a strong binding strength, signifying that artemisinin has the potential to effectively interact with and influence the enzyme’s activity. However, in the case of pyrimethamine, the binding affinity of −8.1 kcal/mol indicates a slightly weaker interaction compared to artemisinin. Nonetheless, this negative value still indicates a favorable binding interaction between the pyrimethamine and the target enzyme.
3.5. Protein–Ligand Interactions of Artemisinin and Pyrimethamine Against SHMT (PDB ID: 1DFO)
The docking score alone provides false-positive results for protein–ligand binding strength and stability [21]. Supporting the docking result by taking into account the key amino acid residues involved in the docking process can make the docking result more reliable and convincing, as well as make it easier to verify the inhibitory activity of the ligands against SHMT. The binding affinities and stable conformations of the ligand within the protein active site are influenced by various factors, including hydrogen bonding, hydrophobic interactions, electrostatic forces, and van der Waals forces between the chemical groups of the ligand and residues at the active site of the protein. In the docked complexes, the interactions at the active site were particularly notable for acidic and basic amino acid residues, which play crucial roles in the activity of SHMT (1FDO) when compared with control that is the standard malaria drug that interferes with the folate metabolic pathway of the parasite. In artemisinin-1FDO complex, Gly A: 211(3.65) and Val A: 204(4.46) are involved in H-bond interactions, Tyr A: 213(5.59) is involved in carbon–hydrogen interaction while Met A: 201(5.92) and Pro A: 216(6.49) participated in π-alkyl and alkyl interactions. In pyrimethamine-1FDO complex, Asn D: 347(3.40 and 4.25) are involved in H-bond interactions while Leu D: 127(4.93) are found to involved in π–σ interactions, and Pro C: 258(4.13), Leu D: 121(6.15), and Phe C: 257(5.84) are involved in π-alkyl and alkyl interactions. The details of these interactions are summarized in Table 3. The interaction observed between the artemisinin and control contributes to stabilizing the ligands in the binding pocket of the receptor as illustrated in Figures 8 and 9, respectively. Therefore, from the result obtained, it can be deduced that artemisinin can readily inhibit the activity of SHMT, thereby preventing the parasite’s growth and reproduction, potentially leading to the development of effective treatments for malaria.
Table 3: Binding affinity and protein–ligand interactions profile of the targeted protein with Artemisinin and control compound.
| Targeted receptor with pdb ID | Compounds | Binding affinity Kcal/mol | Conventional H-bond interactions | Carbon Hydrogen interactions | π–σ interactions | π-Alkyl & Alkyl interactions |
|---|
| Serine hydroxy Methyltransferase (PDB ID: 1dfo) | Pyrimethamine (Control) | -8.1 | Asn D: 347(3.40 & 4.25) | --- | Leu D: 127(4.93) | Pro C: 258(4.13)
Leu D: 121(6.15)
Phe C: 257(5.84) |
| Artemisinin | -9.1 | Gly A: 211(3.65)
Val A: 204(4.46) | Tyr A: 213(5.59) | --- | Met A: 201(5.92)
Pro A: 216(6.49) |
3.6. Major ADME Properties of the Selected Compounds
The major ADME properties of artemisinin were evaluated and analyzed, along with the control compound, using the SWISSADME online tool (http://www.swissadme.ch/index.php), and the results are presented in Table 4. The results indicate that all the compounds exhibited substantial drug-like properties as per the major parameters evaluated. Generally, orally active medicines delivered via the transcellular route should not have a total polar surface area (TPSA) greater than 120 Å., while poorly absorbed compounds have a TPSA greater than 140 Å. [22,23]. In this study, all the ligands had a TPSA range between 77.82 and 53.99 Å. and hence good at permeating cell membrane [Table 4]. The number of rotatable bonds of all the ligands has a number of rotational bonds (nrotb) within the recommended range (0–2) [Table 4]. A drug compound with an nrotb of ≤10 is more likely to have good bioavailability. All the molecules show good lipophilicity (XLogP3) value using the LogS (ESOL) not higher than 6 as a criterion. The pharmacokinetic properties of all the ligand molecules were predicted and are presented in Figure 10 and Table 4. All the compounds obey Lipinski’s rule of five without violation and have a good bioavailability score of [F > 10%] as shown in Table 4.
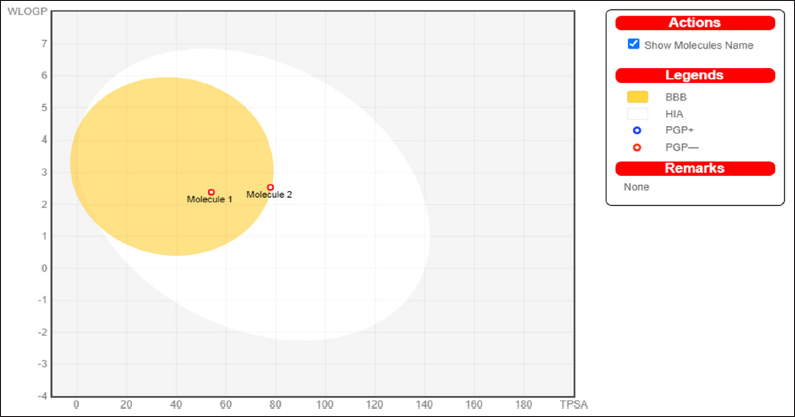 | Figure 10: The boiled egg depicted molecule 1 as a control (pyrimethamine) and molecule 2 as the artemisinin (the investigative compound).
[Click here to view] |
Table 4: Major ADME parameters of the artemisinin and control compounds.
| Molecules | Water Solubility Log S (ESOL) | Lipophilicity Xlogp3 | Polarity TPSA (Å2) | Flexibility rotatable (0<_<9) | GI absorptions | BBB Permeants | P-gp substrate | Lipinski’s violations | Bioavailabity score |
|---|
| Molecule 1 | -3.47 | 2.69 | 77.82 | 2 | High | Yes | No | 0 | 0.55 |
| Molecule 2 | -3.47 | 2.90 | 53.99 | 0 | High | Yes | No | 0 | 0.55 |
3.7. Toxicity Analysis of the Artemisinin Compounds and Control
The toxicity analysis was carried out for artemisinin and control and the findings are presented in Table 5. All the compounds have no AMES toxicity. Moreover, acute oral rat toxicity (LD50), the maximum recommended tolerated dose for a human, hERG I inhibitor, hERG II inhibitor, hepatotoxicity, skin sensitization, and T. Pyriformis, were evaluated, and all the compounds were found to be safe. The details of the toxicity analysis are presented in Table 5.
Table 5: Toxicity analysis of the Artemisinin and control compounds.
Selected
Compounds | AMES toxicity | Max. tolerated human dose (log mg/kg/day | hERG I inhibitor | hERG II inhibitor | LD50
(mol/kg) | LOAEL (log mg/kg | Hepato toxicity bw/day) | Skin sensitization | T.Pyriformi |
|---|
| Molecule 1 | No | 0.113 | No | No | 2.912 | 1.45 | No | No | 0.456 |
| Molecule 2 | Yes | 0.065 | No | No | 2.459 | 1.00 | No | No | 0.322 |
4. CONCLUSION
The study revealed that petroleum ether is the most suitable and efficient solvent for the quantitative and qualitative extraction of artemisinin compound from A. annua L. The findings of the molecular docking analysis further supported the potential inhibitory activity of artemisinin against the SMHT of P. falciparum, thereby inhibiting the DNA and RNA synthesis in the parasite system. This finding is significant for pharmaceutical industries as artemisinin has shown potential effectiveness against the SARS-CoV-2 virus, in addition to its established antimalarial properties.
5. ACKNOWLEDGMENTS
We thank the management of the Herbal Health Research Consortium for valuable support and facilities during the course of this work.
6. AUTHOR CONTRIBUTIONS
Conceptualization was done by Devendra Kumar Pandey. Sample preparation, data collection, and analysis were performed by Ibrahim Isyaku Muhammad and Devendra Kumar Pandey. The first draft of the manuscript was written by Ibrahim Isyaku Muhammad and revised by Devendra Kumar Pandey. All authors read and approved the final manuscript.
7. FINANCIAL SUPPORT AND SPONSORSHIP
This research received no external funding.
8. CONFLICTS OF INTEREST
This study does not involve experiments on animals or human subjects.
9. ETHICAL APPROVALS
This experiment does not involve the use of animal or human subjects.
10. DATA AVAILABILITY
The authors confirmed that, the data supporting the finding of the present study are available within the article.
11. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
12. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Muhseen ZT, Hameed AR, Al-Bhadly O, Ahmad S, Li G. Natural products for treatment of Plasmodium falciparum malaria: an integrated computational approach. Comput Biol Med. 2021;134:104415. [CrossRef]
2. Pillay P, Maharaj VJ, Smith PJ. Investigating South African plants as a source of new antimalarial drugs. J Ethnopharmacol. 2008;119(3):438–54. [CrossRef]
3. Shibeshi MA, Kifle ZD, Atnafie SA. Antimalarial drug resistance and novel targets for antimalarial drug discovery. Infect Drug Resist. 2020;13:4047–60. [CrossRef]
4. Ajah PO, Eteng MU. Phytochemical screening and histopathological effects of single acute dose administration of Artemisia annua L. on testes and ovaries of Wistar rats. African J Biochem Res. 2010;4(7):179–85.
5. Misra H, Mehta D, Mehta BK, Jain DC. Extraction of artemisinin, an active antimalarial phytopharmaceutical from dried leaves of Artemisia annua L., using microwaves and a validated HPTLC-visible method for its quantitative determination. Chromatogr Res Int. 2014;2014:1–11. [CrossRef]
6. Douglas NM, Anstey NM, Angus BJ, Nosten F, Price RN. Artemisinin combination therapy for vivax malaria. The Lancet infectious diseases. 2010 Jun 1;10(6):405–16.
7. Bhakuni RS, Jain DC, Sharma RP, Kumar S. Secondary metabolites of Artemisia annua and their biological activity. Curr Sci. 2001;80(1):35–48.
8. Oh S, Kim BJ, Singh NP, Lai H, Sasaki T. Synthesis and anti-cancer activity of covalent conjugates of artemisinin and a transferrin-receptor targeting peptide. Cancer Lett. 2009;274(1):33–9. [CrossRef]
9. Beekman AC, Wierenga PK, Woerdenbag HJ, Van Uden W, Pras N, Konings AW, et al. Artemisinin-derived sesquiterpene lactones as potential antitumour compounds: cytotoxic action against bone marrow and tumour cells. Planta Med. 1998;64(7):615–9. [CrossRef]
10. Efferth T. Beyond malaria: the inhibition of viruses by artemisinin-type compounds. Biotechnol Adv. 2018;36(6):1730–7. [CrossRef]
11. Kronenberger T, Lindner J, Meissner KA, Zimbres FM, Coronado MA, Sauer FM, et al. Vitamin B6-dependent enzymes in the human malaria parasite Plasmodium falciparum: a druggable target? Biomed Res. Int. 2014;2014:108516. [CrossRef]
12. Nzila A, Ward SA, Marsh K, Sims PF, Hyde JE. Comparative folate metabolism in humans and malaria parasites (part I): pointers for malaria treatment from cancer chemotherapy. Trends Parasitol. 2005;21(6):292–8. [CrossRef]
13. Feng Z, Chen M, Xue Y, Liang T, Chen H, Zhou Y, et al. MCCS: a novel recognition pattern-based method for fast track discovery of anti-SARS-CoV-2 drugs. Brief Bioinform. 2021;22(2):946–62. [CrossRef]
14. Verma AK, Srivastava VK, Srivastava SK. Identification of corticosteroids as potential inhibitor against glycolytic enzyme hexokinase II role in cancer glycolysis pathway: a molecular docking study. Vegetos. 2023;36(1):173–180. [CrossRef]
15. Verma AK, Bala HA, Muhammad II, Muhammad A, Kori AR, Barik M. Virtual screening, molecular docking, pharmacokinetic, physicochemical and medicinal properties of potential curcumin derivatives against Sars-Cov-2 main protease (Mpro). Asian J Pharm Anal Med Chem. 2020;8(4):153–79. [CrossRef]
16. Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. [CrossRef]
17. Pires DE, Blundell TL, Ascher DB. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem. 2015;58(9):4066–72. [CrossRef]
18. Babacan Ü, Cengiz MF, Bouali M, Tongur T, Mutlu SS, Gülmez E. Determination, solvent extraction, and purification of artemisinin from Artemisia annua L. J Appl Res Med Aromat Plants. 2022;28:100363. [CrossRef]
19. Cao J, Yang M, Cao F, Wang J, Su E. Well-designed hydrophobic deep eutectic solvents as green and efficient media for the extraction of artemisinin from Artemisia annua leaves. ACS Sustain Chem Eng. 2017;5(4):3270–8. [CrossRef]
20. Kundu S, Das A, Ghosh B. Optimized extraction of artemisinin from Artemisia annua L. and corroborated quantitative analysis using high-performance thin-layer chromatography. J Planar Chromatogr Mod TLC. 2016;29(5):341–6. [CrossRef]
21. Cheng J, Hao Y, Shi Q, Hou G, Wang Y, Wang Y, et al. Discovery of novel Chinese medicine compounds targeting 3CL protease by virtual screening and molecular dynamics simulation. Molecules. 2023;28(3):937. [CrossRef]
22. Zagórska A, Jaromin A. Perspectives for new and more efficient multifunctional ligands for Alzheimer’s disease therapy. Molecules. 2020;25(15):3337. [CrossRef]
23. De La Nuez A, Rodríguez R. Current methodology for the assessment of ADME-Tox properties on drug candidate molecules. Biotecnol Appl.2008;25(2):97–110.